An Overview of the Central Dogma of Molecular Biology
- Ella Kline
- Sep 22, 2020
- 10 min read
Updated: Jul 16, 2023
Welcome to the central dogma of molecular biology, the one concept that is essential for all life on Earth and is incredibly important when studying biochemistry!
Genes contain all the information that is needed to give rise to life, but this genetic information needs to be maintained and converted into proteins that organisms can use.
In this article, we will discuss three main processes - DNA replication, transcription (which means the conversion of DNA to mRNA) and translation (when mRNA is converted to proteins). These three processes make up the central dogma of molecular biology.
Using regulatory and editing/splicing mechanisms, more information can be found in a single piece of DNA than its nucleotide sequence would suggest.

Figure 1: This shows the central dogma processes that take place as we progress from DNA to proteins. The amount of information obtained from a single gene can be greatly increased through processes such as alternative splicing.
DNA, or deoxyribonucleic acid, is the main form of genetic material found in eukaryotes (e.g. animals and plants) and prokaryotes (e.g. bacteria). It is essential for growth, replication and repair in every cell, thereby allowing for information about an organism to be passed on from one generation to another.
DNA is made of antiparallel strands of deoxynucleotides (also known simply as nucleotides) twisted into a double helix, both of which must be copied during replication. Antiparallel means that one strand of DNA is read in one direction (5’ to 3’), and the other strand is read in the other direction (3’ to 5’).
The nucleotides in each individual strand are joined together by phosphodiester bonds, a type of covalent bond. In every nucleotide, there is a nitrogenous base, a phosphate group and a 5-carbon deoxyribose sugar. In each nucleotide of DNA, the 5-carbon sugar and the phosphate group remain the same, whilst the nitrogenous base determines the ‘letter’ of the genetic code at a particular position.
Besides that, there is Watson-Crick base pairing between opposite nucleotides within the double helix. Watson-Crick base pairing occurs through hydrogen bonding. It is always between adenines (A) and thymines (T) on opposite strands. or between cytosines (C) and guanines (G). These make up the genetic code which allows the storage and processing of information. whereby three adjacent nucleotides make up a codon, and one codon codes for a single amino acid.
Due to Watson-Crick base pairing (A-T and C-G), we can say that the two strands of DNA are complementary to each other. Complementary refers to the ability of two nucleotides to form a perfect base pair with each other, thereby allowing the formation of the double helix of DNA.
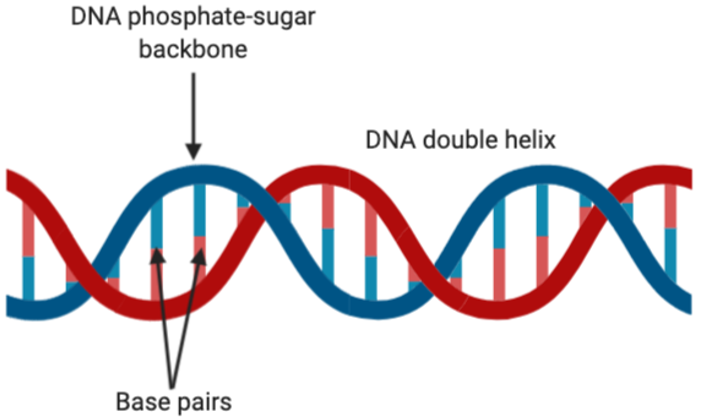
Figure 2: The double helix structure of DNA. This figure shows the base pairing through sticks, where adenine-thymine and guanine-cytosine are always being matched up as a result of Watson-Crick base pairing. Figure created on BioRender.

Depending on the type of organism, the DNA will then be packaged in order to compact it. The purpose of this is to ensure that the DNA will fit into the cell, thereby protecting from external damages.
In eukaryotic cells, DNA is wrapped around histones and is stored in the nucleus. Histones are basic proteins which have DNA wrapped around them to form nucleosomes. Many nucleosomes come together to form chromosomes, and this is how DNA is stored in eukaryotic cells.
On the other hand, in prokaryotes, DNA exists as plasmids, which are circular structures of DNA in the cytoplasm because prokaryotic cells do not have nuclei.
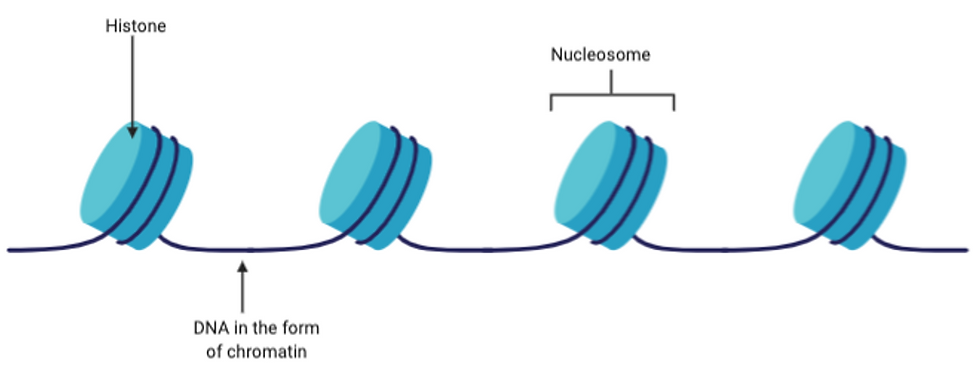

Figure 4: Top diagram showing how DNA is wrapped around the basic histone proteins to form nucleosomes. On the other hand, the bottom diagram shows a plasmid, which is a double stranded, circular DNA that is contained in the cytoplasm of prokaryotic cells. Figure created on BioRender.
Due to these qualities that DNA possess, it is generally a stable structure and so will remain in a cell over its lifetime. If there is any damage or severe mutation(s), the cell would likely die so that it does not negatively affect the whole organism.
DNA Replication (DNA —> DNA)
During DNA replication, there are key enzymes which are used to aid the stabilisation and progression of DNA replication at each stage.
To begin with, DNA polymerases are the main family of enzymes present in DNA replication. They are responsible for the production of new strands of DNA. How exactly are they involved? In essence, DNA polymerases catalyse the formation of the phosphodiester bonds between adjacent nucleotides during the elongation stage of replication. Some also have activity that allows them to repair mistakes during DNA replication.
Other than that, DNA helicases disrupt the Watson-Crick base pairing in DNA and unwind the double helix, therefore giving other enzymes access to the DNA. On the other hand, DNA topoisomerases are a class of enzymes that help convert between the different states of DNA. ‘Different states’ refers to the level of supercoiling of a DNA molecule and whether the DNA is circular or not. In particular, supercoiling is used to compact DNA for storage purposes. In this case, they are relevant because as the helicase unwinds the double helix, the topoisomerase (also known as DNA gyrase) relieves stress in order to prevent the DNA from breaking.
DNA replication is a semi-conservative process, meaning that the new DNA contains one old (parental) strand and one newly-synthesised (daughter) strand. This was experimentally confirmed by Meselon and Stahl, after it was hypothesised by Nikolai Koltsov. Since DNA is antiparallel and its replication is bidirectional, the parental strands can thus be split into the leading and lagging strands. The leading strand is unwound in the 3’ to 5’ direction, allowing elongation in the 5’ to 3’ direction. The lagging strand has an antiparallel direction, and so it is unwound in the 5’ to 3’ direction.
The first stage of replication begins at the origin, an adenine-thymine rich section of DNA, where the DNA helicase will unwind the double-stranded DNA. At the same time, DNA topoisomerase acts just ahead of the replication fork. This produces single-stranded DNA in the structure of a replication fork, with the leading and lagging strands separated. Single-stranded binding proteins will then coat the unwounded DNA to stabilise the strands and prevent reformation of double-stranded DNA.

Figure 5: The formation of the replication fork with single-stranded binding proteins coating the single-stranded, unwounded DNA on both the lagging and leading strands. DNA helicase and DNA topoisomerase are shown as ovals located ahead of the replication fork. These unwind the double-stranded DNA to allow access to other enzymes, such as DNA polymerase. This figure was adapted from Bacterial Replication Fork: Synthesis of Lagging Strand (2010). Figure created on BioRender.
All known DNA polymerases can only carry out elongation, they would be adding nucleotides in the 5’ to 3’ direction only. Thus, in areas where the lagging strand is acting as the template, replication must be initiated many times as the new strand must be synthesised in the 3’ to 5’ direction.
This is carried out by a series of enzymes including DNA primase, which creates RNA primers of short sequences of nucleotides, known as Okazaki fragments, that are added to the lagging strand. Moving forward, DNA ligase then fills in the gaps between these fragments. DNA helicase and DNA polymerase will form a complex with DNA, allowing the replication of both the leading and lagging strands simultaneously. Also, beta-clamps are present in this complex, and they stabilise the DNA.
All these may sound very complicated and overwhelming at first. But don’t worry, try checking out this video, which may be useful to strengthen your understanding on how all these machineries work together: https://youtu.be/HSoKeKV5ecs?t=154.
Transcription (DNA —> RNA)
RNA (ribonucleic acid) is another type of nucleic acid besides DNA. RNA often exists as a single strand and is usually far more transient (meaning lasts for a short time) than DNA.
RNA contains nucleotides, which similarly to DNA, are joined together by phosphodiester bonds. However, unlike DNA, RNA contains a nucleotide called uracil (U) and does not contain thymine (T), so uracil (U) will pair with adenine (A) instead of thymine (T).
Types of RNA
There are three key RNAs you need to know about: mRNA (and pre-mRNA), tRNA and rRNA.
Messenger RNA, or mRNA, is produced directly from DNA during transcription and is used to convey genetic information to aid the production of proteins in translation. In eukaryotes, pre-mRNA is produced directly from transcription and parts of this must be removed to produce mRNA.
Transfer RNA, or tRNA, is used in translation to carry amino acids to the ribosome. tRNAs have a unique structure, so that they are folded to create a specific shape that allows an amino acid attached to one end and the other end possesses an anticodon that will hydrogen bond with a codon of mRNA. So they act as an adaptor between mRNA and proteins.
Ribosomal RNA, or rRNA, along with proteins make up ribosomes which carry out translation.
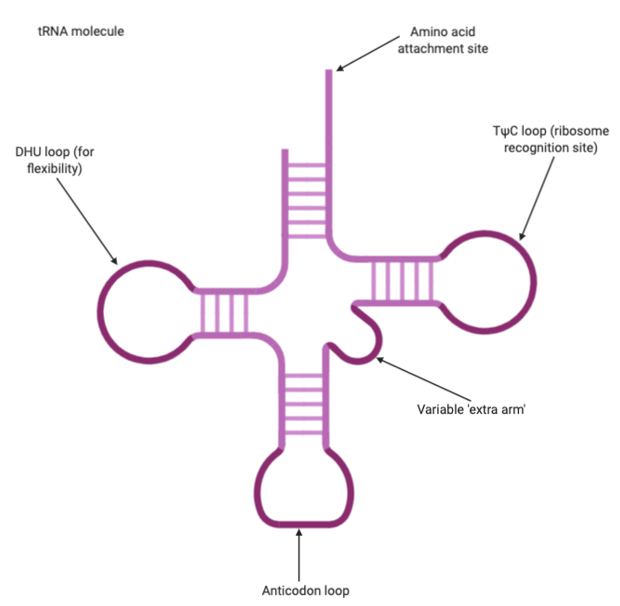
Figure 6: tRNA molecule with the key structures labelled – the amino acid attachment site, the anticodon loop and other sites that allow for flexibility and recognition by the ribosome. Figure created on BioRender.
Transcription uses RNA polymerase to create RNA transcripts from DNA. The DNA is separated into a coding strand and a template strand that is complementary to the coding strand. Following that, the RNA polymerase will add nucleotides to the template strand to produce a transcript that has the same ‘code’ as the coding strand.
The RNA polymerase will interact with a particular sequence of DNA called the promoter, thereby signalling for transcription to begin. In this sequence, 12-15 base pairs are unwounded to form a transcription bubble, and transcription can begin. The RNA polymerase is able to move along the DNA, using the Watson-Crick base pairing rules to add nucleotides to the template strand of DNA. Eventually, the RNA polymerase will reach a terminator sequence of DNA, which signals for the end of transcription, causing the release of the DNA and RNA transcript.
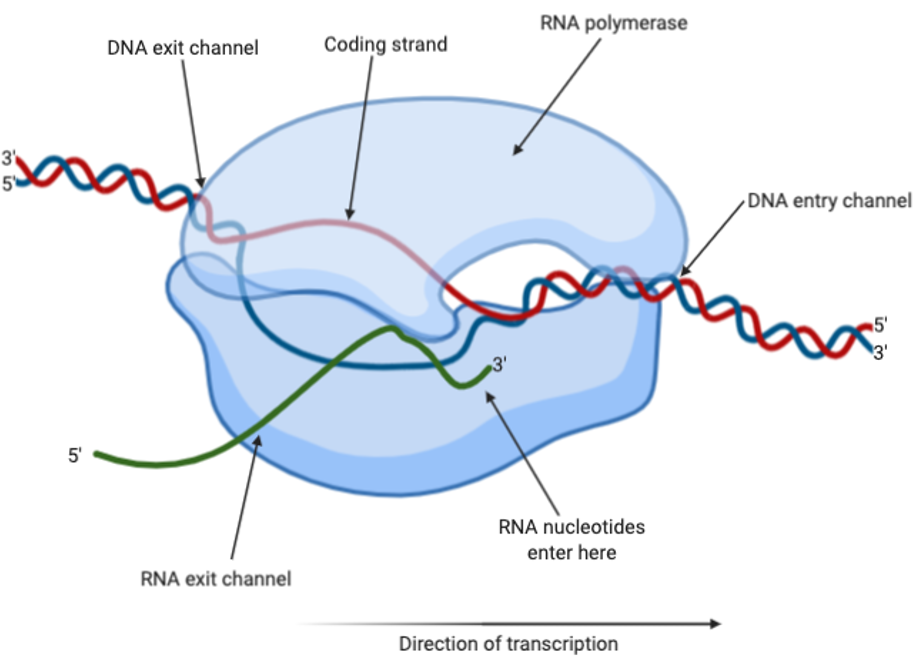
Figure 7: RNA polymerase carrying out transcription with DNA and RNA. The coding strand is shown in red, whereas the template strand is shown in blue and the RNA is shown in green. Figure created on BioRender.
In prokaryotes, there is one type of RNA polymerase which has a highly conserved 3D structure. This means that the overall structure does not differ much, if at all, between different organisms. It has five subunits that all play slightly different roles during transcription.
Whereas in eukaryotes, there are three types of RNA polymerase. However, we are most concerned with RNA polymerase I, which produces pre-mRNA transcripts in the nucleus. In particular, RNA polymerase I is unable to start transcription without help from other proteins in the nucleus, so it is slightly more complicated than RNA polymerase in prokaryotes (though it still follows a similar principle).
Splicing
Pre-mRNA contains exons, which are coding pieces of mRNA, as well as introns, which do not contain coding sequences. Since introns do not have any coding sequences, they must be removed before the mRNA transcript is transported out of the nucleus. This process is known as splicing.
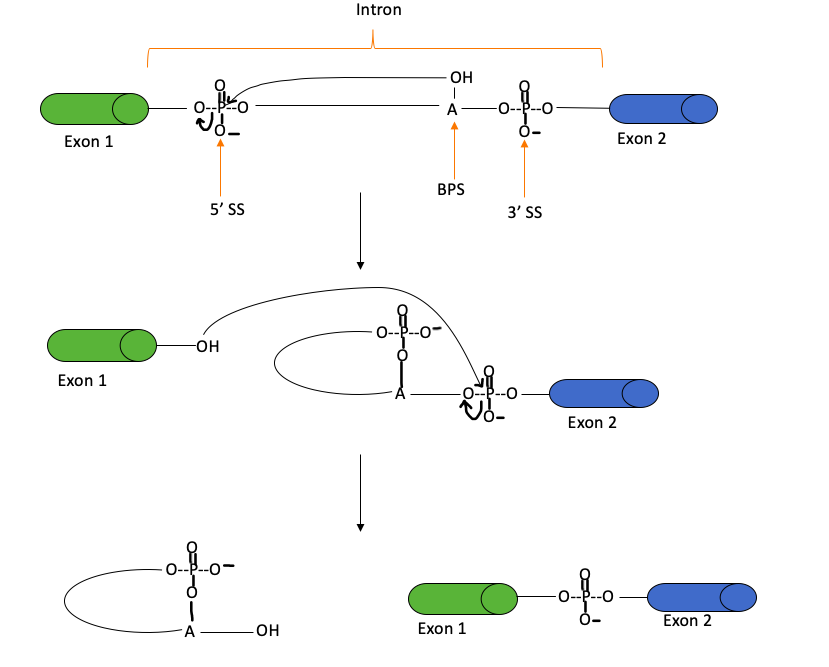
Figure 8: The splicing mechanism, which occurs through two transesterification reactions, to remove introns and produce mRNA. A transesterification reaction involves the formation of one ester from another through the exchange of an alkoxy moiety. During splicing, the 2’-hydroxyl group of the SS attacks the 5’ SS, in the second reaction, the liberated hydroxyl group attacks the 3’ SS to produce mRNA and a lariat (i.e. a looped structure). “SS” stands for the splice site, whereas BPS represents the branch site. This figure was adapted from Clancy (2008).
Translation (RNA —> Proteins)
Translation occurs after transcription, and the process takes place within the ribosomes inside a cell.
Interestingly, there are 64 possible codons, but only 20 amino acids. This makes the genetic code redundant because there are multiple codons that lead to the production of the same amino acid.
Meanwhile, one codon will code for a start codon (AUG), which signals for the start of translation. More specifically, the first amino acid produced from translation is generally methionine.
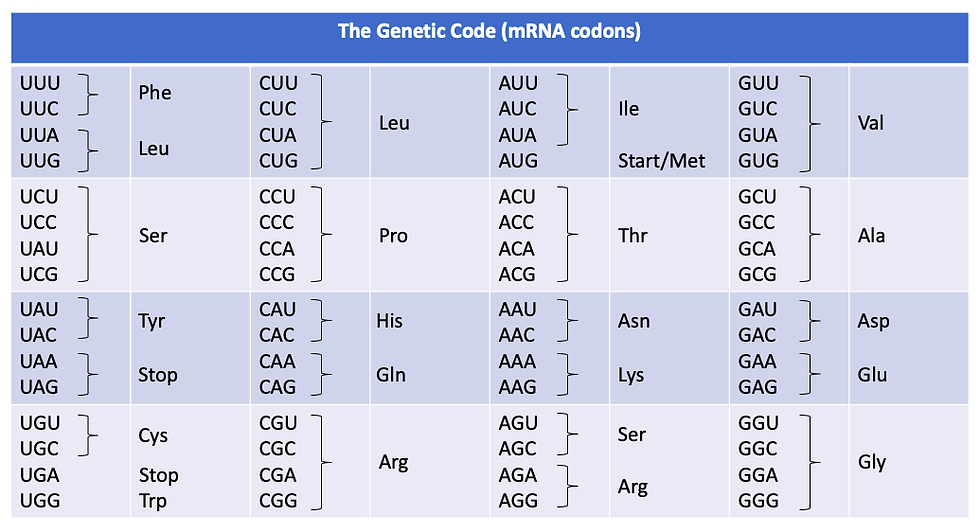
Figure 9: Table showing the genetic code, which translates mRNA codons into amino acids.
Ribosomes have two subunits, a small and large one, with an mRNA-binding site and three tRNA-binding sites (E stands for the exit site; P represents the peptidyl-tRNA site; A signifies the aminoacyl-tRNA site). Eukaryotes have 80S ribosomes, made up of a 60S large subunit and a 40S small subunit. On the other hand, prokaryotes have 70S ribosomes with a 50S large subunit and a 30S small subunit. ‘S’ refers to the sedimentation coefficient in Svedberg units, which measures how fast they sediment; this is related to the mass, density and shape of the ribosomes.
Similar to DNA replication, translation can be split into initiation, elongation and termination. In prokaryotes, the small subunit is able to recognise the start of the mRNA at a sequence of mRNA called the Shine-Dalgarno sequence. In both prokaryotes and eukaryotes, initiation factors aid recognition and the loading of mRNA onto the ribosomes. These include IF1 and IF2 (IF = initiation factor), which play different roles in the initiation of translation. IF1 binds to the A site of the small subunit of the ribosome to produce a conformational change to aid initiation of translation. Meanwhile, IF2 binds to allow more favourable binding of the first tRNA.

Figure 10: Simplified diagram of a ribosome with the large and small subunits labelled. As well as this, the E-, P- and A-sites are labelled, and they each bind a tRNA molecule. The E- (or exit) site is where the tRNA will leave the ribosome; the P- (or peptidyl-tRNA) site is where the tRNA is bound to the polypeptide chain that is being made; and the A- (or aminoacyl-tRNA) site is where a tRNA molecule first binds to the ribosome. The mRNA loading site is also shown, whereby during translation, the mRNA molecule fits into a groove that is only present when the small and large subunits are connected. Figure created on BioRender.
During the elongation stage, the ribosome will be bound to the mRNA with three adjacent codons aligned to the E, P and A sites, but there are generally only two tRNAs in a single ribosome. The two amino acids attached to the two adjacent tRNA molecules will form a peptide bond. The ribosome will shift down the mRNA molecule three nucleotides (or one codon) at a time until the stop codon is reached. Once multiple amino acids are attached, they form a polypeptide chain, which can then be processed further to form a protein. The small and large subunits of the ribosome will then dissociate, freeing the mRNA and ribosome.
One more thing, it is important to note that the same mRNA can be used for translation over and over again until it is degraded.
Key definitions
The central dogma allows the transfer of information from the genes of an organism into usable information in the form of proteins. These processes can be complex and require different cell machinery to carry them out.
Central dogma: The transfer of information from genetic material to proteins that the cell can use. It is widely accepted that the flow of information is from DNA —> RNA —> proteins
DNA replication: The semi-conservative replication of DNA. This process occurs so that daughter cells will have the correct amount of genetic information. It requires enzymes to catalyse several processes in order to ensure its accuracy and efficiency
Transcription: The process of producing mRNA transcripts from DNA, which can then be taken to the ribosome so that it could be translated into proteins
Translation: mRNA codons temporarily bond with tRNA anticodons to allow amino acids, specifically attached to tRNAs, to create a polypeptide chain
Extra resources
These videos may be especially useful to visualise DNA replication/structure, transcription and translation:
DNA replication: https://www.youtube.com/watch?v=8kK2zwjRV0M
DNA replication, transcription and translation: https://www.youtube.com/watch?v=6gUY5NoX1Lk&list=PLF3iv2SQQbj9oA0ScwkrDqKbw0NE6h_w9&index=6
Transcription and translation: https://www.youtube.com/watch?v=bKIpDtJdK8Q
Author: Ella Kline, BSc Biochemistry
Disclaimer: All figures created using BioRender are intended solely for educational purposes and not for profit.
