COVID Diagnostics: What’s The Deal With Real-Time RT-PCR?
- Devyani Saini

- Feb 23, 2021
- 11 min read
Updated: Jul 6, 2023
Now that we are more than a year into the global pandemic caused by the novel coronavirus SARS-CoV-2, the term “real-time reverse transcription polymerase chain reaction” (RT-PCR) has become commonplace in mainstream journalism as the primary diagnostic method for the deadly virus.
Large-scale coronavirus testing has played a huge role in influencing the policy of major governments on controlling and containing the virus. Here, we will get into the finer biochemical details of the real-time RT-PCR method for detecting the SARS-CoV-2 as well as a look into how it was developed.
Polymerase chain reaction – the discovery that started it all
In 1983, Kary Mullis and his colleagues started developing a method to amplify a small sample DNA using a pair of primers (short DNA sequences of around 20 nucleotide base pairs) at either end of the desired sequence along with the addition of DNA polymerase (the enzyme involved in DNA replication).
The only drawback back then was that the DNA polymerase enzyme would get denatured at the high temperature (around 95°C) required to separate the double-stranded DNA (dsDNA) and allow the primers to anneal. Nonetheless, this problem was solved by using the DNA polymerase from the organism Thermophilus aquaticus (Taq), which was stable at high temperatures. The resultant method was eventually termed the “polymerase chain reaction”.
How do you perform one?
Here’s a little more detail on how it works...
A typical reaction mixture for PCR will have the following components:
The DNA template, which is the sample containing the sequence that we want to amplify
DNA primers, or more specifically, the forward and reverse primers. The forward primer is complementary to the start of the sequence (the 5’ end), while the reverse primer is complementary to the end of the sequence (3’ end). In essence, they both help the DNA polymerase to have a ‘starting point’ from which it extends the rest of the DNA strand
The DNA polymerase enzyme
A buffer solution, which keeps the enzymes and DNA at the optimal pH
The deoxynucleoside triphosphates (dNTPs). These are the nascent bases (A, T, G, and C) in which the DNA polymerase adds to the newly developing DNA strand
In particular, the DNA polymerase, buffer, and dNTPs are usually combined into a “Master Mix”, which can be purchased from molecular biology suppliers such as New England Biolabs or Thermo Fisher to make the preparation of the reactions a lot easier.
Meanwhile, the machine used to perform a PCR is called a thermal cycler, which is named so because they run through a series of temperature changes required for the PCR to amplify the quantity of DNA.
A typical temperature cycle for a PCR could look like this:

Figure 1: A simplified illustration of how the three-step PCR works. The first step is melting – in which the double-stranded DNA template unwinds into single strands at around 95°C to 98°C. Then, the primers anneal (join) at their respective complementary sequences at around 60°C. After that, the next step is elongation, whereby the DNA polymerase synthesizes a new DNA strand using the dNTPs in the PCR mixture.
All in all, these three steps constitute one cycle, and a typical PCR lasts around 40 cycles of amplification. In this manner, up to billions of copies of DNA can be generated for use in experiments and analyses. After the PCR, a PCR purification step can be carried out to separate the DNA sample from any residual DNA and other unwanted components.
In essence, the development of the PCR has revolutionized molecular biology and opened the doors to a huge variety of techniques, including the RT-PCR method, which is now used to detect SARS-CoV-2. In fact, this discovery was deemed to be so impactful to the world of science that it was awarded the Nobel Prize in Chemistry in 1993!
The PCR method is critical for COVID-19 testing, but it requires a few additional things to detect the viral genetic material, one of which is reverse transcriptase, which will be discussed in the next section.
Reverse transcriptase – the next step
In 1970, David Baltimore, Howard Temin, and Satoshi Mizutani discovered the presence of RNA-dependent DNA polymerases in the virions (form of the virus outside the host cell) of RNA viruses. To be more specific, Temin and Mizutani discovered it in Rous sarcoma virions, whereas Baltimore found out about it in murine leukemia virus. In particular, these enzymes were observed to convert viral RNA into DNA so that the virus could integrate itself into the host genome. Upon observing this, the scientists then hypothesized that this characteristic could be exploited in molecular biology.
And turns out you can.
RNA-dependent DNA polymerases, more commonly called “reverse transcriptases”, are capable of synthesizing complementary DNA (cDNA) from an RNA template in contrast to the central dogma of molecular biology, which states that RNA is produced from a DNA template (Figure 2).
After that, the cDNA obtained from using reverse transcriptase would later be amplified using PCR – hence the term RT-PCR, which stands for “reverse transcription polymerase chain reaction”.

Figure 2: The central dogma of molecular biology. The central dogma states that DNA is transcribed to RNA, which is then translated into protein sequences. However, reverse transcriptases go against the central dogma as they allow RNA to be converted back to DNA (cDNA, more specifically).
In short, RT-PCR has many applications in molecular biology. One of them would be monitoring the mRNA levels in a cell in response to different conditions (such as a change in temperature or pH levels). Meanwhile, another example would be detecting foreign RNA such as SARS-CoV-2 within a patient sample.
But, to get a result that can be ruled as ‘positive’ or ‘negative’ for the virus, there is one additional step in the diagnostic process…
Quantitative real-time PCR – bringing it all together
Quantitative real-time PCR, shortened to qPCR, is a modification to PCR which allows the amount of DNA being produced during each cycle to be monitored in real-time (hence its name).
This method was first demonstrated by Russel Higuchi and his team in an experiment using a dye called ethidium bromide (EtBr), which is a compound that binds to dsDNA. They monitored PCR cycles using a video camera to capture the fluorescence emitted by EtBR when it binds to the dsDNA produced from the PCR. This experiment thus showed that it was possible to look at a PCR in real time and to see when exactly does the DNA start to amplify exponentially (as in at which cycle number of the PCR).
They later demonstrated that the initial number of DNA copies was correlated to the number of cycles it took for fluorescence to be detected. This is, therefore, the quantitative nature of qPCR because it allows us to see how much DNA there is in a sample.
A qPCR graph commonly looks like this:

Figure 3: An example of how the result of a qPCR may look like. The key data point is the cycle threshold (Ct) value, which tells us the amount of RNA present in the sample. It can be defined as the cycle number where the fluorescence of the sample exceeds that of the background and becomes detectable. In essence, the lower the Ct value, the higher the amount of RNA. And in contrast, the higher the Ct value, the lower the amount of RNA. Not to mention, an important note is that the Ct values obtained from different qPCR machines or from different qPCR reagents cannot be compared to each other due to the differences in sample processing methods.
Using real-time RT-PCR to detect SARS-CoV-2
Real-time RT-PCR (often shortened to rRT-PCR) combines qPCR and reverse transcription PCR together to quantify the amount of mRNA in a sample. This is the method that is used to detect the SARS-CoV-2 coronavirus from nasal and throat swab samples.
Firstly, the viral RNA is extracted from the sample and converted into cDNA using reverse transcriptase. Then, the qPCR reaction mixture is set up in a PCR tube, with forward and reverse primers and the Master Mix added to it. In addition to that, the qPCR Master Mix contains the fluorescent dye (e.g. SYBR Green, SYBR Safe, ethidium bromide) required to monitor the PCR cycles. The reaction mixture is then placed into a qPCR machine, which differs slightly from a regular thermal cycler (it has additional features suited to qPCR data processing). Meanwhile, a control reaction is also set up alongside the sample, whereby it does not contain any viral RNA in it.
The machine then cycles through the melting, annealing, and elongation temperatures - as in a normal PCR - a total of 40 times. Overall, the entire process is expected to take around two hours.
Finally, the data is extracted in the form of Ct values, which tells us if the sample has a significant amount of viral RNA, and thus whether the sample is ‘positive’ for SARS-CoV-2 or not. As a general rule-of-thumb, if the Ct value is lower than 40, it is usually deemed as a positive result. However, this may differ from lab to lab depending on which qPCR reagents and thermal cycler machines are used.
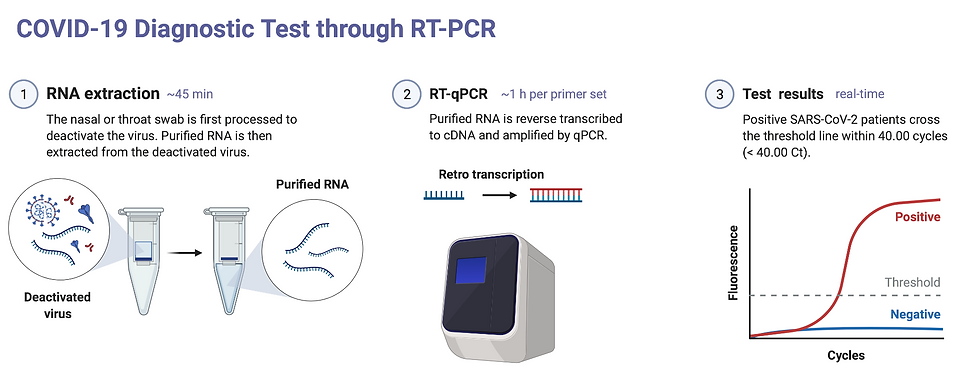
Figure 4: A schematic diagram illustrating the COVID-19 diagnostic process by real-time RT-PCR. (1) RNA extraction is usually done using a commercial kit from companies such as Qiagen. (2) The purified RNA is reverse transcribed to cDNA and then amplified using real-time PCR (qPCR). (3) After the qPCR runs, the result is obtained in graphical form where the Ct value indicates whether the sample is positive or negative for SARS-CoV-2.
Evaluating rRT-PCR as a diagnostic method - is it actually effective?
As we saw in the previous section, the Ct values tell us about whether a sample contains the SARS-CoV-2 novel coronavirus. Like all diagnostic methods, it has advantages and disadvantages, which we will be exploring in this section.
The advantages of rRT-PCR
The main advantage of real-time RT-PCR for detecting SARS-CoV-2 is its extreme accuracy and specificity. Accuracy means that it is able to measure what it is supposed to measure - i.e. it only measures specific sections of the SARS-CoV-2 genome. In fact, the accuracy of RT-PCR can be attributed to the primers that are chosen. This is because the primers flank the sequence of the viral genome, ensuring that only the relevant sections are amplified during the PCR protocol (see Figure 1).
On the other hand, specificity refers to how many people who are not infected actually test negative (a.k.a. the “true negative rate”). From collating the results of several studies, an overall specificity rate of 95% was determined for the rRT-PCR used to detect SARS-CoV-2. This means that if 100 people were tested to not have COVID-19 using the rRT-PCR technique, of these, 5 people may actually be tested positive (even though it is confirmed that they do not have it). These, in turn, are called false positives. In essence, the higher the specificity, the lower the chance for false-positive results. Nonetheless, the good news is that RT-PCR has a very high specificity, which basically means that false positives are unlikely.
The limitations of rRT-PCR
Despite having high specificity, rRT-PCR has only moderate sensitivity. Sensitivity refers to how many people who are infected actually test positive (a.k.a. the “true positive rate”).
On that note, it is estimated that the test has a 70% sensitivity rate from a review of the literature. This means that if 100 people have COVID-19, 30 people may return with a negative result. This is termed as a false-negative and is arguably far more dangerous than a false positive because the person may think they do not have COVID-19, but is in fact passing it on to others.
Additionally, the test is only taken at one point in time, so it can’t really tell us how the disease is going to progress, or at what stage of the disease a person is at. This essentially could be problematic because the exact time when the test is taken in the timeline of the disease can also affect the Ct value, and hence our interpretations of it (see Figure 5).
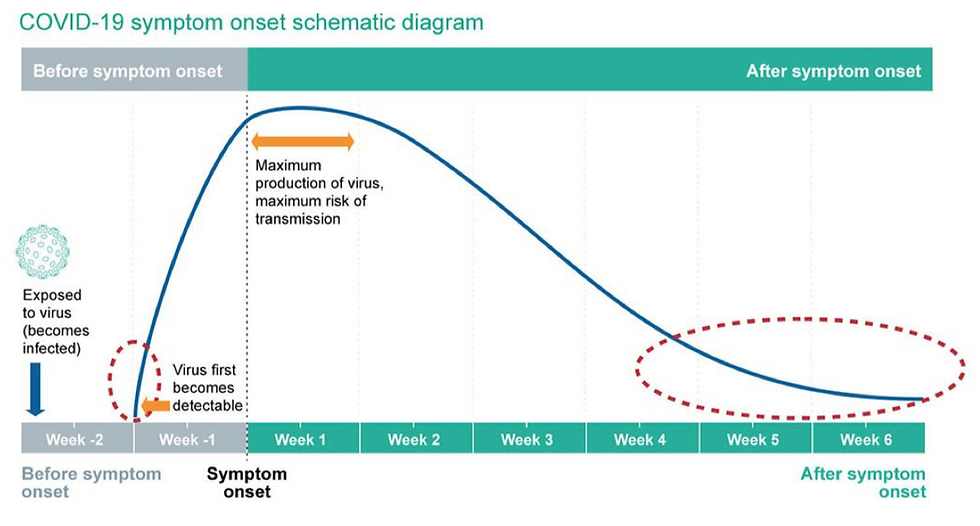
Figure 5: The above figure from Public Health England outlines the timeline of disease, from infection point to eventual recovery. The blue line indicates the viral load in arbitrary units. Areas indicated by the red dashed circles represent the time periods in which the positive rRT-PCR test results may have a high Ct value, and hence indicate a low viral load.
Taking these limitations into account, should a positive test have a high Ct value, we won’t know if the person has just been infected or if their immune system has begun clearing the virus (hence resulting in a lower viral load and higher Ct). Thus, it is essential that Ct values are interpreted in tandem with clinical information and patient history.
Other than that, the rRT-PCR test has a few other drawbacks. For instance, it is time-consuming, expensive, and requires laboratory expertise to perform. To give you a better idea, an rRT-PCR test from a private lab in the UK costs upwards of £100! This takes into account the cost of qPCR reagents, running the machine, paying laboratory staff, as well as a profit margin (well, it is a private lab after all). Additionally, as the samples require careful handling and processing, the whole process can take upwards of four hours if we were to combine all the steps required to obtain a result (See Figure 4).
Other tests used to detect SARS-CoV-2
Fortunately enough, we do have options.
For instance, another commonly used test to detect SARS-CoV-2 is the rapid antigen test or lateral flow device (LFD) test. This test uses immobilized antibodies that correspond to viral antigens (biological markers on the virus) which produce an easy-to-read result in a similar way to a pregnancy test. Besides that, these tests are very cheap and easy to use (in fact, no specialized training is needed!). However, they don’t have the accuracy of rRT-PCR.

Figure 6: Lateral flow device tests provided by the UK Government. Negative results will not have any SARS-CoV-2 antigens, and hence the antibodies immobilised on the strip will not bind to anything and there will be no visible colour change. On the other hand, positive results will show a colour change. In particular, the intensity of the colour corresponds to the viral load - a darker band means the person is more infectious. Void results occur when the LFD test is faulty or if it has been used incorrectly.
The main advantage of rapid antigen tests is that they have a high specificity - a positive antigen test would almost always mean the patient will have a positive rRT-PCR test (though not always the other way around). This is because rapid antigen tests require a higher viral load to yield a positive result as compared to rRT-PCR. As a result, rapid antigen tests can often yield false-negatives. And on that note, the lower sensitivity would be the main disadvantage of the LFD test as people with lower viral loads could go undetected by it but may still be capable of infecting others.
So… what’s next?
The real-time RT-PCR test is so far the most accurate and most specific test currently available to detect SARS-CoV-2. It has a few limitations nonetheless, but when used in tandem with the LFD tests, it is an effective tool in the fight against the COVID-19 pandemic.
While government testing labs utilise the RT-PCR test, businesses, schools, and universities are opting for the cheaper and simpler LFD tests. Nevertheless, using both these tests in tandem will be the most optimal way to monitor the virus, as they can cover for each others’ weaknesses. For example, patients discharged from hospital after being treated for COVID-19 often test positive for months afterward (which is usually due to the presence of inactive traces of the virus within their system). By using a rapid antigen test, it can be confirmed that the patient is non-infectious, and hence not a danger to public health.
All in all, in this article, we have outlined the biochemical background of the tests used to detect the coronavirus, but this is still ever-changing.
In fact, as you are reading this, many private laboratories are developing new and improved versions of both the rRT-PCR and LFD tests. One such example is the ‘FRANKD’ (Fast Reliable Accurate Nucleid acid-based Kit for COVID-19 Detection) marketed by GeneMe. This testing kit utilises reverse transcription loop-mediated isothermal amplification, a nucleic acid amplification method that requires only one step and one constant temperature. Who knows - this could potentially revolutionise how the National Health Service (NHS) tests for COVID-19, leading to even faster, more widespread, and more efficient mass testing!
Author
Devyani Saini
MRes Molecular & Cellular Biosciences
Imperial College London
