Introduction
Viral hepatitis is a serious condition caused by a hepatitis virus infection. It causes the inflammation (i.e. swelling) of our liver, resulting in a potentially fatal damage of this organ. So far, five hepatitis viruses have been discovered - hepatitis A, B, C, D, and E.
The name ‘hepatitis’ refers to the fact that the virus’ primary infection site is hepatocytes (i.e. liver cells), but does not reflect its evolutionary relationship, meaning that the hepatitis viruses are not related and each of them actually come from a different family.
The hepatitis C virus (HCV) is a blood-borne virus which was discovered in 1989 following a large number of mysterious post-transfusion cases. Since then, the scientific community has made huge progress in understanding the virus. Tests are now available to screen blood and prevent transfusion-associated hepatitis, and antiviral drugs capable of curing over 90% of infected individuals have been developed. However, in 2015, 71 million people were still living with chronic HCV infection. And in 2016, the virus was estimated to have killed 399,000 people. Taking those into account, HCV is clearly a huge global health issue and there is still lots more work to be done if we are to control it.
In this article, we will be exploring the biology of HCV, the nature of the disease it causes, and the story behind its identification and discovery, which won three scientists the 2020 Nobel Prize in physiology or medicine. I would recommend reading the ‘Introduction to Virology’ article first, if you haven’t done so already, to help you understand some of the concepts in this one.
About the virus
Classification
HCV is the only member of the genus Hepacivirus, which is in the family Flaviviridae. Another genus of this family is flavivirus, which includes yellow fever virus and dengue fever virus.
HCV can be further divided into six genotypes - the genomes of these different genotypes have some variations, known as sequence divergence. This variation is due to the high mutation rate of the virus. Genotype 1 and 2 viruses are found worldwide, whilst genotypes 3 to 6 have their own restricted geographic ranges. For example, genotype 3 is found in India, Europe and the United States. On the other hand, genotype 5 is mainly restricted to South Africa, whilst genotype 6 is distributed across various countries in South East Asia.
Meanwhile, the disease manifestations are generally the same for each genotype, although some research suggests those infected by HCV genotype 3 may develop liver cancer sooner. In contrast, the antiviral therapy offered to patients is dependent on the genotype they are infected with.
Genome
HCV has a positive sense, single-stranded RNA genome, which means it can be translated directly by the host’s ribosome. The genome consists of a single large open reading frame (ORF; a sequence that isn’t interrupted by a stop codon). This ORF is translated by host ribosomes into a long polyprotein of around 3000 amino acids. This polyprotein contains three structural proteins and seven non-structural proteins as seen in Figure 1.
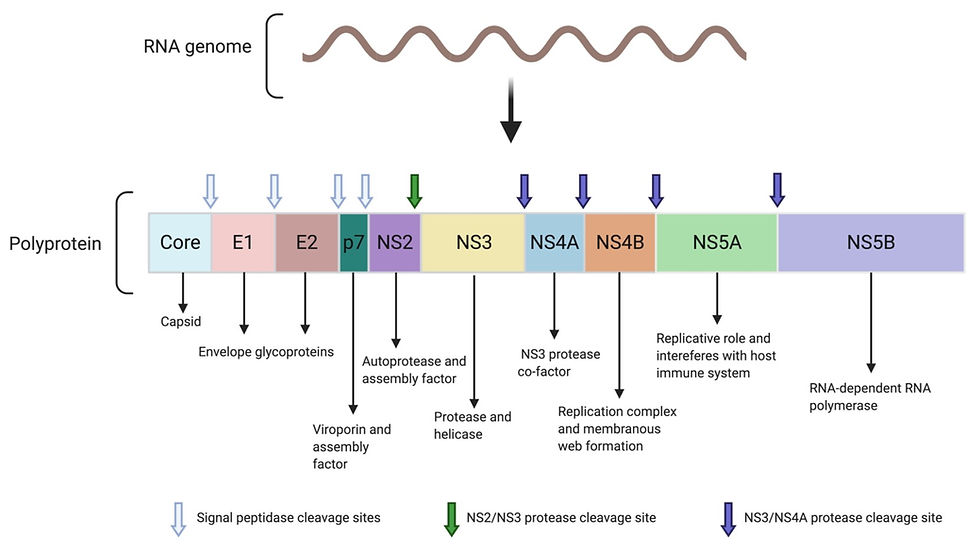
Figure 1: The proteins encoded by the HCV genome. The single-stranded RNA genome of hepatitis C is transcribed and translated into a single polyprotein. The polyprotein is then cleaved into individual, functional proteins by either the host cellular signal peptidase, NS2/NS3 protease or NS3/NS4A protease, as indicated on the figure. The main functions of the cleavage products are also shown. This figure was adapted from Abdel-Hakeem (2014), Protective immunity against hepatitis C: many shades of gray and created using BioRender.
The first structural protein is the core protein. This is an RNA binding protein which forms the viral capsid. The capsid is a protein shell which surrounds and protects the viral RNA genome.
The other structural proteins are E1 and E2 (called envelope 1 and 2, respectively). These two glycoproteins are embedded in the host-derived lipid membrane, where they interact tightly with each other to form the E1-E2 heterodimer. They are involved in the attachment and entry of the virus into the host cell.
On the other hand, p7 is a small, non-structural transmembrane protein which isn’t part of the viral particle. But instead, it forms a hexamer ion channel known as a viroporin in host cell membranes, where it can help with viral assembly. However, its exact role hasn’t yet been elucidated.
NS2 (non-structural protein 2) acts as a protease (i.e. an enzyme that catalyses the breakdown of proteins). In particular, it has a protease domain which works together with a domain found on NS3 (non-structural protein 3), as seen in Figure 2a. Together, the NS2-NS3 protease cleaves the junction between NS2 and NS3 in the polyprotein, freeing the individual proteins. After this self-cleavage, the NS2 loses its protease activity and is also involved in coordinating the assembly of new viral particles in the infected host cell.
NS3 has two major domains, enabling it to carry out a variety of functions. Firstly, it forms a protease with NS2, as mentioned previously. However, it also functions independently as a protease which cleaves the polyprotein to release individual non-structural proteins. It also uses its protease activity to cleave host proteins involved in the innate immune response. This is the initial, non-specific immune response to HCV infection which involves detection of the virus and, in some cases, clearance of it. By interfering with this immune response, HCV can suppress the immune response in the early stages of its infection.
NS3 needs to interact with its cofactor, NS4A, in order for it to function as a protease. This interaction is illustrated in Figure 2b. Finally, NS3 acts as a helicase – an enzyme which unwinds viral RNA so it can be replicated.
The NS4B protein sits in the host endoplasmic reticulum and forms the replication complex (which replicates the viral RNA genome) along with various other non-structural proteins. It is one of the viral components needed for the formation of the membranous web - a region in the cytoplasm where viral replication takes place. We will come back to the membranous web and its significance later on.
NS5A has an important role in viral RNA replication. It also interacts with host proteins to interfere with cell signalling pathways, such as those needed for cell growth. Meanwhile, NS5B is the RNA-dependent RNA polymerase (RdRp). This enzyme is essential for RNA replication to synthesise new viral genomes for new virion particles.
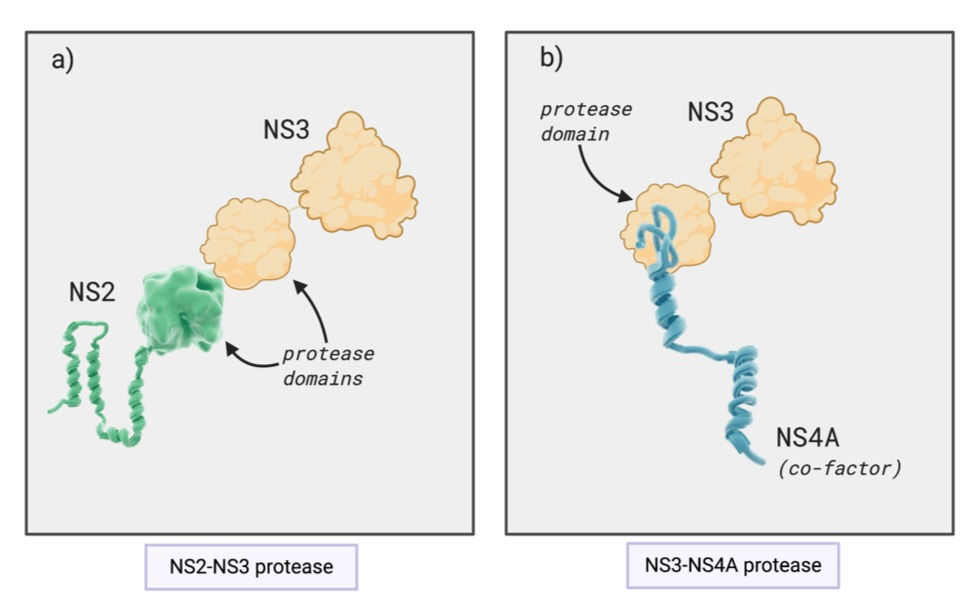
Figure 2: Interactions between viral proteins to form proteases. (a) The protease domains of NS2 (non-structural protein 2) and NS3 together form a protease which cleaves the junction between NS2 and NS3 in the polyprotein. (b) NS3, specifically its protease domain, interacts with its cofactor NS4a. This protein complex cleaves the polyprotein to release individual non-structural proteins. This figure was adapted from Hepatitis C online (2020), HCV proteins and was created on BioRender.
The stages of the viral life cycle are illustrated in Figure 3.
1. Cell attachment
The first stage of infection involves binding to the receptor on the target cell. The initial attachment of HCV to the host cell is mediated by binding to two receptors: the low-density lipoprotein receptor (LDLr) and heparan sulphate proteoglycans (HSPGs). After this, E1 of the E1-E2 heterodimer in the viral envelope binds to the receptor SR-B1 and CD81.
In general, a virus’s receptor determines its tropism (the cells/tissues a virus can infect). Given the receptors required for its cell entry, hepatocytes (liver cells) are the main tropism of HCV.
2. Endocytosis
Once the viral particle has attached to its host cell, it is internalised into the cell in a process called endocytosis, where the host cell membrane surrounds the virus and folds inwards to form an endosome.
3. Fusion
The endosome, which contains the viral particle, has an acidic pH which triggers fusion between the host endosomal and membrane. This fusion, mediated by E2, allows the capsid to ‘uncoat’, thereby releasing the genomic viral RNA into the cytoplasm.
4. Translation
The viral RNA is translated directly by the host ribosome into a polyprotein, which then undergoes ‘proteolytic processing’ by cellular and viral proteases to cleave it into single proteins.
Firstly, the signal peptide peptidase (i.e. a cellular enzyme which catalyses the breakdown of proteins) cleaves the core, E1, E2 and p7 proteins. Then, the NS2 protein and NS3 protease domain cleave NS2 from NS3. Finally, the NS3-NS4A membrane complex cleaves the rest of the non-structural proteins.
5. RNA replication
HCV, like a range of other single-stranded RNA viruses, is capable of rearranging host cell membranes to form a membranous web. This is a collection of vesicles where the viral RdRp replicates the genome, producing multiple copies to be packaged into new viral particles.
Replicating in these vesicles is a clever way for the virus to evade immune detection. The innate immune system has receptors in the cytoplasm that recognise pathogen-associated molecules which aren’t found in mammalian cells. For example, double-stranded RNA, an intermediate of viral replication (see Figure 4), is not normally found in the cell cytoplasm. Therefore, its presence indicates pathogen invasion. By restricting replication to a membranous web, the virus is able to hide its dsRNA from specific cytoplasmic and thus prevent the host innate immune response being triggered.
6. Assembly
The exact mechanism of HCV assembly is not completely understood yet.
What we know so far is that it involves core proteins coming together and assembling into the capsid, which surrounds newly synthesised genomic RNA. Viral assembly is thought to take place in the endoplasmic reticulum. The E1 and E2 proteins are present in the rough endoplasmic reticulum membranes - so when the virus buds from the endoplasmic reticulum, it acquires a lipid envelope with the envelope proteins embedded in it. The new virion particle is finally released from the host cell by exocytosis, where it goes on to infect new cells or be transmitted to a new host.

Figure 3: Hepatitis C life cycle. The virus binds to its target cell using the LDLr (low density lipoprotein receptor), HSPGs (heparan sulphate proteoglycans), SR-B1, or CD81 [1]. The virus is then endocytosed into the cell [2]. pH-dependent fusion occurs between the endosomal and viral membrane, allowing the RNA genome to be uncoated and released into the cytosol [3]. The genome is directly translated [4] by host ribosomes to produce a polyprotein, which is cleaved by host and viral proteases. The RdRp (RNA-dependent RNA polymerase) replicates the viral genome in the membranous web, which is made up of DMVs (double membrane vesicles) [5]. In the endoplasmic reticulum, core proteins surround the genome and the viral particle is assembled [6]. This new virus then matures in the Golgi apparatus and is released from the cell [7]. This figure was adapted from Grassi et al. (2016), Hepatitis C virus relies on lipoproteins for its life cycle and was created using BioRender.
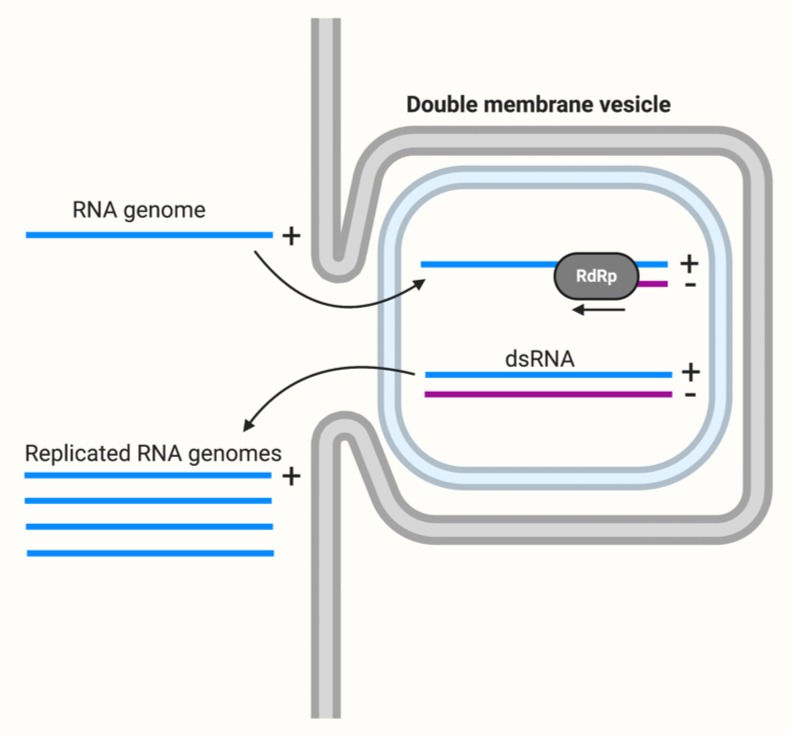
Figure 4: Replication of the RNA genome in a double-membrane vesicle. The positive sense (+), single-stranded RNA genome is replicated by the RdRp (RNA-dependent RNA polymerase) to produce a complementary negative sense (-) strand. The double-stranded RNA (dsRNA) molecule present at this stage is hidden from cytoplasmic receptors. And finally, the negative sense strand is used as a template to create genome copies. This figure was adapted from ViralZone, Positive stranded RNA virus replication and created using BioRender.
HCV and disease
HCV can cause acute or chronic infection.
Around 30% of infections are acute. This means that any symptoms will occur soon after infection, although 80% of individuals are asymptomatic following initial infection. These acute infections tend to be cleared spontaneously within six months.
However, the remaining 70% who are chronically infected. Chronic viral infections are persistent and can even remain for life and tend to have long incubation periods. These chronically HCV-infected individuals can develop life-threatening conditions, mainly liver cancer and cirrhosis (a liver disease caused by long-term liver damage). In fact, the risk of developing cirrhosis within 20 years ranges from 15 to 30%. The stages of HCV infection are summarised in Figure 5.
Transmission
HCV is a blood-borne virus. The main methods of transmission are sharing needles for injecting drugs, unscreened blood transfusions and reusing medical equipment without proper sterilisation. The virus can be transmitted vertically from an infected mother to baby, although this is less common.
Diagnosis
A serological assay which detects anti-HCV antibodies (i.e. antibodies specific to HCV) can identify people who have been infected by the virus. If these antibodies are present, a nucleic acid test to detect the RNA of HCV is then carried out to confirm the individual is chronically infected. This is because someone who has been infected in the past but cleared the virus by a strong immune response will still test positive for anti-HCV antibodies but negative for HCV RNA.
Cancer
Epidemiological evidence indicates that persistent HCV infection is major risk factor for development of hepatocellular carcinoma (HCC). The latency period between the initial infection and development of cancer is very long, often lasting decades. The exact mechanism by which the virus causes cancer has not been determined yet, but it is currently thought that the consequences of HCV infection, such as cell death and proliferation, chronic inflammation and cirrhosis are responsible.
Chronic inflammation is mediated by the continuous activation of inflammatory cells, which release harmful substances such as reactive oxygen and nitrogen species (ROS and RNS, respectively). These trigger oxidative DNA damage, creating an environment favourable to tumorigenesis (meaning the formation of a tumour) (Check out Figure 5). This is one possible explanation for the link between chronic inflammation and cancer.
The virus itself also induces change in cells which can result in transformation (this is when cells become malignant and begin to divide uncontrollably and spread).
For example, it can impair DNA repair pathways, increase proliferation and decrease apoptosis (programmed cell death, used to kill cells proliferating at abnormal rate or with DNA damage). HCV proteins can also inhibit tumour suppressor genes, which as the name suggests, are critical in preventing cell transformation. The combination of chronic inflammation and HCV infection therefore increases cellular DNA damage whilst also decreasing its ability to repair the damage or kill the potentially cancerous cell (See Figure 5).
The fact that only a relatively small proportion of HCV-infected individuals actually develop HCC suggest that environmental factors also contribute to the risk of carcinogenesis (i.e. the development of cancer).
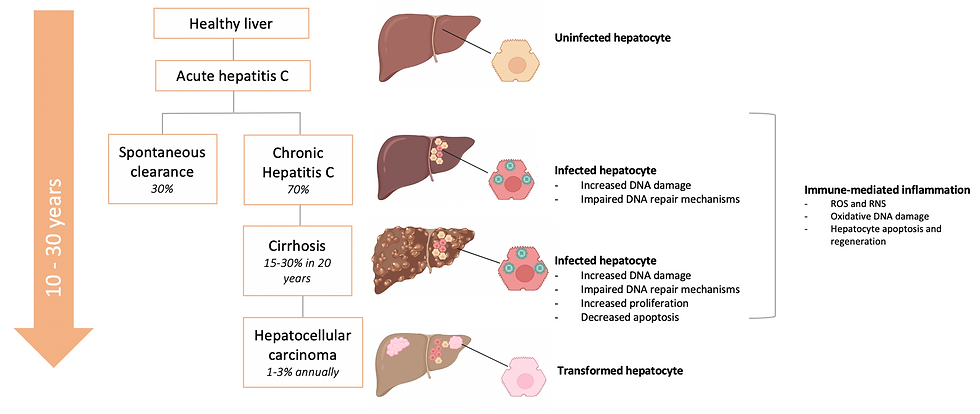
Figure 5: The stages of hepatitis C virus (HCV) infection. Infection can lead to hepatocellular carcinoma indirectly by host-mediated inflammation, or directly by HCV-mediated mechanisms. HCV can interfere in a range processes in infected cells to create a pro-tumorigenic environment, potentially leading to cell transformation and carcinogenesis (cancer formation). This figure was adapted from Mitchell et al. (2015), How do persistent infections with hepatitis C virus cause liver cancer? and Holmes et al. (2013), Hepatitis C.
Treatment
Acute HCV is rarely diagnosed, so treatment at this stage is uncommon and sometimes unnecessary as the immune response can clear the infection.
Treatment with drugs, known as Directly Acting Antivirals (DAAs), is available for chronic infection and clears the virus in at least 90% of patients.
DAAs target different stages of the HCV lifecycle. For example, NS3/4A protease inhibitors are a class of DAAs which target the virus’ NS3/4A protease enzyme. Other classes include NS5B inhibitors and NS5A (RNA-dependent RNA polymerase) inhibitors. Drugs from different classes are often taken in combination to improve the treatment efficacy and to reduce the risk of the virus evolving resistance against the drugs.
A vaccine for HCV is not yet available, as there are a number of significant challenges hindering development efforts. One problem is the genetic heterogeneity of HCV - the various genotypes and subtypes of the virus can have significantly different sequences in genes for some of their proteins. This means a vaccine against one strain may not be protective against another. Also, HCV could quickly evolve to escape the long-term immunity induced by the vaccine. Another challenge is our limited understanding of how exactly HCV becomes chronic and what type of immunity is required to be protected against HCV infection (this is known as the correlate of immunity).
Discovery of HCV
In the 1960s, the hepatitis B virus was determined to be the cause of blood-borne hepatitis. Once the causative agent was identified, tests for screening blood were introduced to reduce the number of hepatitis cases in people receiving blood transfusions.
Despite this screening, Harvey J. Alter at the United States National Institute of Health noticed that there was still a large number of transfusion-associated hepatitis cases. Tests for the hepatitis A virus, which is spread through direct contact or contaminated food or water, showed that this virus was not responsible.
As many as 10% of transfusion cases were resulting in what became known as ‘non-A, non-B viral hepatitis’ (NANBH). In 1978, Alter transferred plasma (the liquid component of blood) from NANBH patients to chimpanzees who subsequently developed the disease. This showed that the NANBH was caused by an infectious agent, whilst further studies indicated the pathogen was a virus.
After that, it took over a decade for this novel virus to be identified, whereby…
Qui-Lim Choo, George Kuo and Michael Houghton and their colleagues collected nucleic acids from the blood of infected chimpanzees to create a collection of DNA fragments. The next step for the group was to determine which fragments, if any, belonged to the mystery virus. However, this proved too challenging as the majority of fragments were derived from the chimpanzee genome. Instead, they used RNA from the infected chimpanzees to generate another cDNA collection. This cDNA was inserted into bacterial cells, which were left to grow into colonies (i.e. a solid group of cells, all derived from a single parent). As the cells grew, they produced proteins encoded by the cDNA fragments. See Figure 6 for a summary of this process. The group hoped that at least one of these proteins would be viral. Serum (blood with clotting factors removed) from NANBH patients was then used to try and ascertain whether any of these proteins belonged to the mystery virus.
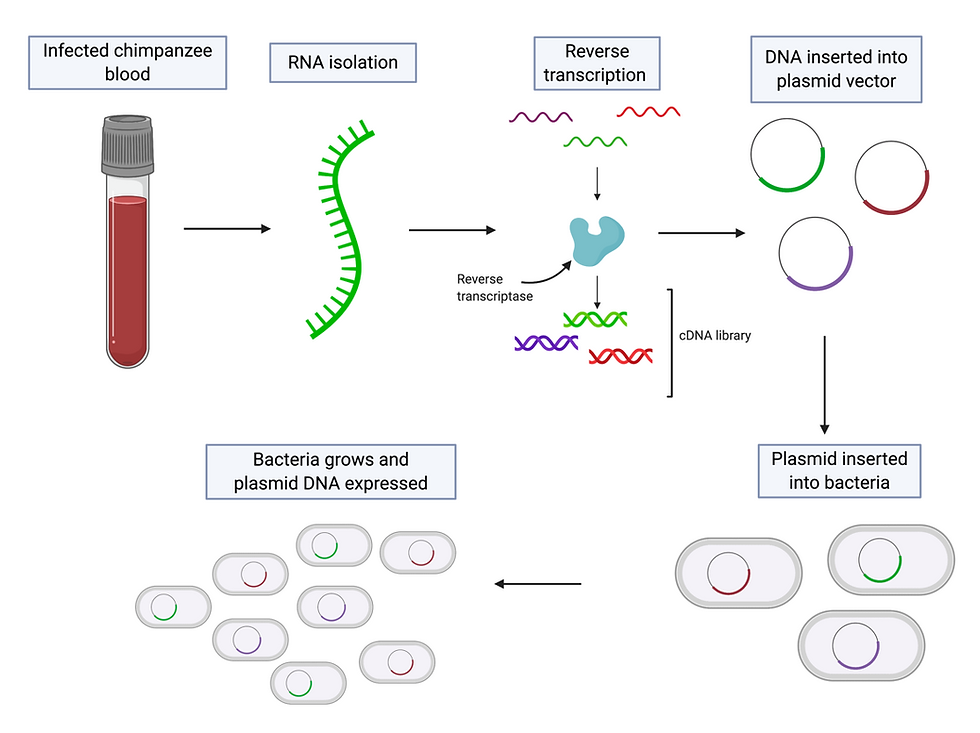
Figure 6: cDNA formation and expression. RNA is isolated from the blood of infected chimpanzees. Reverse transcription is carried out by reverse transcriptase - this is where the isolated RNA is used as a template to produce complementary DNA. The cDNA is then inserted into a vector, such as a plasmid (a circular DNA strand molecule found in the cytoplasm of bacteria). The plasmid is then added to bacteria. As the bacteria grows, it expresses the DNA on the plasmid vector and produces the protein it encoded by it. This figure was adapted from GOLDBIO, Reverse Transcriptase & cDNA Overview & Applications and created using BioRender.
The blood of these patients should have antibodies against the virus, and these anti-HCV antibodies would bind to complementary viral proteins and therefore show which colonies contained cDNA derived from viral RNA. This method was successful - the team screened one million bacterial colonies and found one which produced viral proteins. This new virus was named hepatitis C virus, and the results were published in 1989.
One HCV had been isolated, a blood test was developed and used to screen blood donations for the virus. This massively improved the safety of blood transfusions and led to a huge reduction in the number of post-transfusion hepatitis cases.
However, it still needed to be proven that the virus alone caused hepatitis. Charles M. Rice injected the virus isolated by Houghton, Choo and Kuo into chimpanzees, but the virus failed to replicate and cause infection. Rice suspected that the novel virus had a very high mutation rate, like most RNA viruses, so as it replicated it generated numerous mutations which accumulated in the genome. This led to the formation of variants which were incapable of replicating and causing disease.
Rice then used genetic engineering to create a new HCV variant that was less likely to contain these detrimental mutations. This variant was then injected into chimpanzees, and they eventually developed clinical symptoms similar to those in humans with NANBH with the virus found in their blood. As described in the 1997 paper published by Rice and his colleagues, this was a solid evidence showcasing that the virus alone was causing these mysterious transfusion-associated hepatitis cases.
Fast forward to today, Alter, Houghton and Rice jointly received the 2020 Nobel Prize in Physiology or Medicine for discovering HCV.
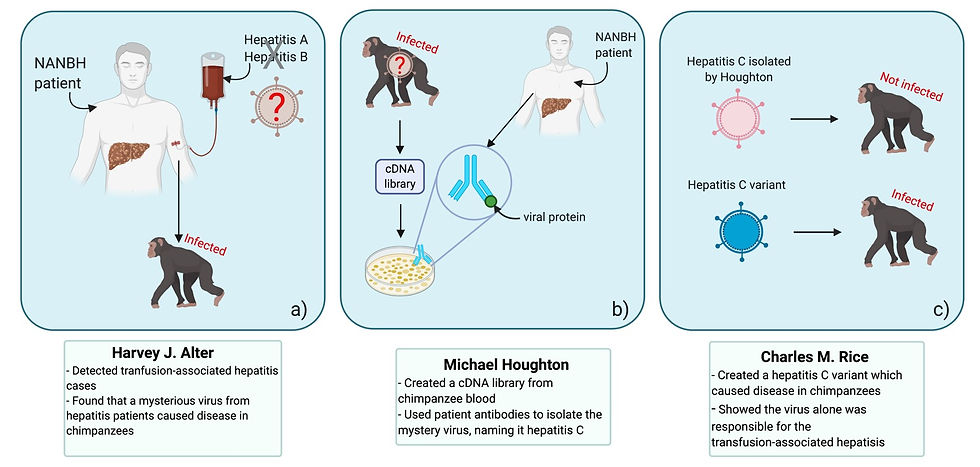
Figure 7: The findings of Harvey J. Alter, Michael Houghton and Charles M. Rice, the winners of the 2020 Nobel Prize for Physiology or Medicine, were essential in the identification of the hepatitis C virus. In particular, (a) Alter detected a large number of hepatitis cases in blood transfusion recipients. Tests showed that the hepatitis A and B viruses weren’t responsible, so the disease was named NANBH (non-A, non-B hepatitis). Alter transferred plasma from NANBH patients to chimpanzees, who contracted the disease, indicating that the causative agent was infectious. Meanwhile, (b) Houghton created a library cDNA (complementary DNA) fragments derived from RNA present in infected chimpanzee serum. The fragments were inserted into bacteria. Each resulting bacterial colony produced the protein encoded in the cDNA fragment. Antibodies specific to the virus, present in the blood of NANBH patients, bound to the colonies producing viral proteins. This allowed the hepatitis C virus to be isolated. And last but not least, (c) Charles M. Rice showed that the virus isolated by Houghton failed to cause disease in chimpanzees. He then made a variant which was capable of infected chimpanzees, proving that the hepatitis C virus was responsible for the mysterious NANBH cases. This figure was adapted from Compound Interest (2020), The 2020 Nobel Prize in Physiology/Medicine: The discovery of the hepatitis C virus and was created using BioRender.
Why is this important?
As mentioned in previous articles, understanding the biology of a viral pathogen is essential for developing treatments and preventative measures against diseases. For example, elucidating the lifecycle of a virus and the proteins involved allows us to develop drugs targeting different stages. The hepatitis C virus still infects millions of people every year and can cause serious disease and mortality in a significant proportion of cases. It is clear that we need to develop better treatments and controls in order to beat the virus.
The 2020 Nobel Prize awarded to Alter, Houghton and Rice marks a ground-breaking achievement in the fight against this virus. Their innovative work and perseverance are not only inspiring but also essential for the rapid development of blood tests and anti-viral drugs and has undoubtedly saved a great number of lives across the globe.
Author: Ambar Khan, BSc Biological Sciences
Disclaimer: All figures created using BioRender are intended solely for educational purposes and not for profit.

Comments