Introduction to Bioinformatics: What is Bioinformatics? [Part One]
- Adekale Idris
- Dec 10, 2020
- 5 min read
Updated: Jul 15, 2023
Bioinformatics is a field of science at the interface between computer science and biological sciences. Understanding the history of bioinformatics helps us know how bioinformatics came to be and gives us an insight into current bioinformatics research.
A brief history
The foundations of bioinformatics were laid more than 50 years ago when computers were not available, and DNA could not yet be sequenced. In 1965, Margaret Dayhoff undertook what might have been the first major bioinformatics project. She developed the first protein sequence database called "Atlas of Protein Sequence and Structure". She’s regarded as the first bioinformatician. Subsequently, the Brookhaven National Laboratory in the early 1970s established the Protein Data Bank (PDB) for archiving three-dimensional protein structures. At first, this database could only store up to a dozen protein structures as compared to its storage capability now (about 30,000 structures today). Needleman and Wunsch were the first to develop an algorithm for comparing sequences. This development paved the way for the routine sequence comparisons and database searching practised today by modern biologists.
The 1980s were important in the history of bioinformatics due to the start of the Human Genome Project (HGP). HGP provided a major boost for the development of bioinformatics. The development of GenBank and fast database searching algorithms such as FASTA by William Pearson and BLAST by Stephen Altschul and coworkers were also developed in the 1980s. Another milestone in the development of bioinformatics was recorded in the 1990s; the genomes of three important model organisms were published: Haemophilus influenzae, S. cerevisiae, Caenorhabditis elegans. The use of the internet and next-generation sequencing led to an increase in the influx of data and a rapid generation of bioinformatics tools in the 1990s-2000s.
Today, the field of bioinformatics has lots of bioinformaticians, however, there is no definite agreement on the exact definition of a bioinformatician is. Some authors suggested that the term should be for those specialized in the field of bioinformatics, including those who develop and maintain bioinformatics tools. On the other hand, it was also suggested that any user of a bioinformatics tool should be allowed the status of a bioinformatician (Who qualifies to be a bioinformatician?) There are lots of bioinformatics related databases and tools. Bioinformatics is now part of the curriculum of most biological sciences programs. For more on the history of bioinformatics, check out History of Bioinformatics.
What is bioinformatics?
There are lots of definitions available in the literature and on the internet; the only difference in these definitions is that some are more inclusive than others. So, this article will adopt the definition proposed by Luscombe et al (What is Bioinformatics?) in defining bioinformatics as a union of biology and informatics: ‘Bioinformatics involves the technology that uses computers for storage, retrieval, manipulation, and distribution of information related to biological macromolecules such as DNA, RNA, and proteins.’ See figure 1 for the 3D structure of a protein.
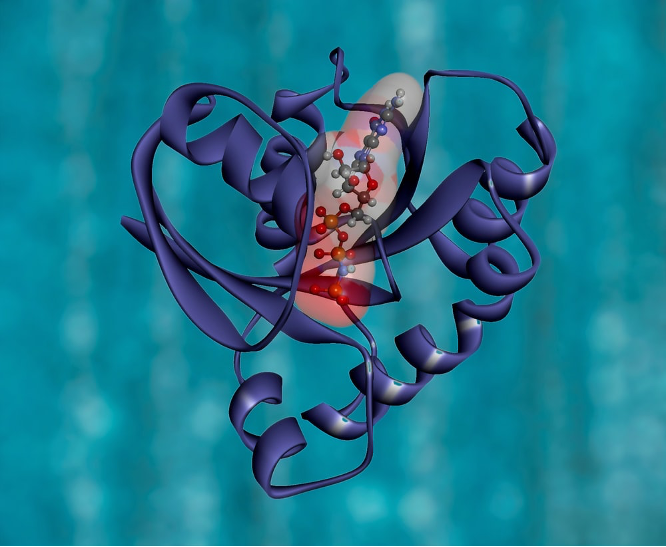
Figure 1: KRAS Protein Structure. RAS is a family of related proteins that is expressed in all animals. KRAS is one of three RAS genes found in humans. RAS genes are mutated in approximately one-third of all human cancers.
Bioinformatics differs from computational biology although they are related. Bioinformatics is considered by some as computational molecular biology which involves sequence, structural, and functional analysis of genes and genomes and their corresponding products. However, computational biology encompasses all biological areas that involve computation. Generally, bioinformatics involves the use of information science and statistics to understand biological macromolecules while computational biology emphasizes the development of theoretical methods, computational simulations, and mathematical modelling.
What are the aims of bioinformatics?
The ultimate goal of bioinformatics is to better understand a living cell and how it functions at the molecular level. Hence, the aims of bioinformatics can be subdivided into three parts:
Bioinformatics organizes data in a way that allows researchers to access existing information and to submit new entries as they are produced
To develop tools and resources that aid in the analysis of data.
To use these tools to analyze the data and interpret the results in a biologically meaningful manner.
What does bioinformatics cover?
The scope of bioinformatics consists of the development of computational tools and databases and their application to analyse biological data in order to better understand living systems. Such analyses often generate new problems and challenges that could lead to the development of new and better computational tools.
There are three aspects of bioinformatics analysis:
Molecular sequence analysis: This has to do with an analysis that involves sequence alignment, sequence database searching, genome comparison, gene and promoter prediction, and phylogeny.
Molecular structural analysis: This has to do with an analysis that involves nucleic acid structure prediction, protein structure prediction, protein structure classification, and protein structure comparison.
Molecular functional analysis: This has to do with an analysis that involves protein interaction prediction, gene expression profiling, etc.
These analyses are dependent on each other and are often used together to produce results. For example, protein structure prediction depends on sequence alignment data; clustering of gene expression profiles requires the use of phylogenetic tree construction methods derived from sequence analysis.
How and where have we used this evolving field?
Bioinformatics has applications in several areas such as drug design, forensic DNA analysis, biotechnology etc. In drug design, understanding the three-dimensional structures of proteins allows the design of molecules capable of binding to the receptor site of a target protein with great affinity and specificity. Computational based studies significantly reduce the time and cost necessary to develop drugs with higher potency. It also allows molecules to be designed with fewer side effects and less toxicity as compared to molecules developed using the traditional trial-and-error approach when designing drugs.

In forensics, results from molecular phylogenetic analysis (this involves using phylogenetic tree construction methods to generate of phylogenetic tree) shows the relationship between biological data) have been accepted as evidence in criminal courts.

In the healthcare sector, high-speed genomic sequencing coupled with sophisticated informatics technology will allow doctors in a clinic to quickly sequence a patient’s genome. It will also allow them to easily detect potential harmful mutations, engage in early diagnosis and effective treatment of diseases.

In agriculture, plant genome databases and gene expression profile analyses (genomic tools in plant breeding) have played an important role in the development of new crop varieties that have higher productivity and more resistance to disease.
Bioinformatics is a powerful science with some drawbacks
Bioinformatics is dependent on the information gathered and the algorithms being used for subsequent analysis. If something is wrong with one or all of these factors, there will be problems with the results. The quality of data and the sophistication of the algorithms affect the quality of bioinformatics predictions. Sequence data from high throughput analysis often contain errors. If the sequences are wrong or annotations incorrect, the results from the downstream analysis will be unreliable. Algorithms that lack the capability and sophistication to truly reflect reality often make incorrect predictions that make no sense when placed in a biological context. For example, errors in sequence alignment can affect the outcome of structural or phylogenetic analysis.
Slow computational rates also affect algorithms, even accurate ones. A choice between accuracy and computational feasibility has to be made. Hence, most of the time, less accurate but faster algorithms have to be used. Caution should always be exercised when interpreting prediction results. It is a good practice to use multiple tools or programs, if available, and perform multiple evaluations. A more accurate prediction can often be obtained if one draws a consensus by comparing results from different algorithms.
Bioinformatics is an evolving field of science that holds great potential in transforming biological research into a more quantitative and predictive research. There is a need to know the various types of databases, the type of information (sequences) stored in these databases, and how to access them.
Keep a lookout for the next article: Introduction to Bioinformatics II: Biological foundations and databases.
Author: Adekale Idris, BSc Biochemistry
