The Cornerstones of Crime Scene Investigations
- Huong Giang Nguyen
- Dec 28, 2020
- 12 min read
Updated: Jul 15, 2023
In April 2003, a decaying body of a mother and her unborn child washed ashore in California, shocking its residents.
DNA testing was used to identify the bodies as they were in an advanced state of decay. They were soon discovered to be the bodies of Laci Peterson and her child, Conner, both of whom were reported missing by her husband.
However, the husband quickly became the primary suspect as direct evidence (Laci’s hair) was found on a pair of pliers on his boat. And it turns out that the suspicions were right - using comparison microscopy, the Federal Bureau of Investigation (FBI) found out that the hair was identical in terms of its pigmentation, medulla, and cuticle when compared to a hair sample known to belong to Laci.
All in all, this case demonstrates how criminals may overlook even the tiniest details like hair and the fibres of clothes, and these minute hints can serve as important leads for crime scene investigations thanks to the various technological advances that we have.
In this article, we will explore some of the techniques that are used in forensic analysis, and have a brief discussion on the future directions of this field.
The intriguing power of mass spectrometry
Mass spectrometry (MS) is an experimental technique used to identify molecular ions based on their mass-to-charge (m/z) ratio.
The process first begins with the samples being vaporized and then converted into charged ions by an ion source. They then move on to the mass analyzer, where the ions are deflected by a magnetic field at varying degrees depending on their mass-to-charge ratio. In general, the lighter and more highly charged the ions are, the greater their deflection will be. These different ions are then detected (by a detector) and a mass spectrum is produced.
To keep things simple, you can almost equate the mass spectrum to some sort of stick diagram, with each peak corresponding to ions of different m/z ratios. Or to be a little more specific, the mass spectrum is a graph that showcases the relative abundance of ions with reference to their m/z ratios.
Given that each compound has a unique mass spectrum, we can compare the mass spectrum obtained during our experiment to the existing spectral libraries (of known compounds) in order to identify the mystery substance under investigation.
Gas chromatography-coupled mass spectrometry (GC-MS) is useful for identifying inflammable compounds
In many cases, chromatography would be carried out before mass spectrometry analysis in order to improve the separation of the constituents within a mixture. This is particularly useful when analysing compounds that possess similar chemical properties and weight (e.g. the police may need to differentiate between similar gasoline brands when investigating a hit-and-run case). This, in turn, would reduce the frequency of experimental errors.
Gas chromatography (GC) is a separation method used to distinguish between samples that readily evaporate at room temperature (e.g. gases or organic molecules).
The process first begins with liquid samples being vaporized via heating. Constituents of the mixture are then separated based on their different binding affinities within the separation column. These different binding affinities thereby correspond to the varying times needed for the different molecules to travel to the end of the column (see Figure 1).
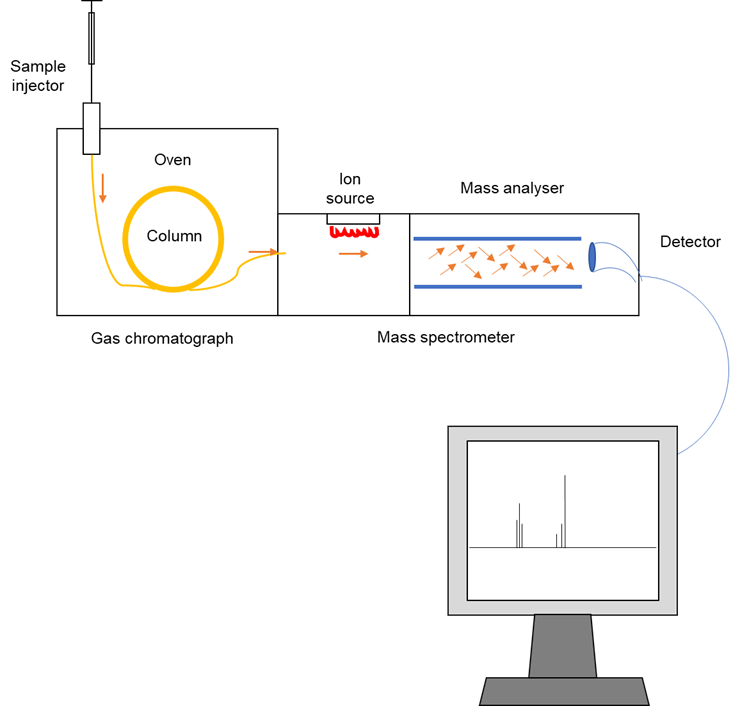
Figure 1: An overview of the gas chromatography-coupled mass spectrometry (GC-MS) workflow. The sample is applied onto a column (usually arranged in a long coil) consisting of a stationary and mobile phase. The stationary phase is usually a polar liquid that coats the column and interacts strongly with other polar compounds. On the other hand, the mobile phase is usually inert (unreactive). In general, components that interact with the stationary phase with greater affinity tend to exit the column later; whereas less polar compounds travel faster and hence exit the column sooner. As a result, the components of the mystery mixture leave the column at different times. These are then directed into the ion source of a mass spectrometer, where the compounds are converted into ions (i.e. ionized) by an electron beam. Following that, the newly generated ions are identified based on their mass-to-charge ratios, with the results displayed as a mass spectrum on the computer. The data obtained can then be used to piece together the chemical structure of the compound in question when the researchers conduct a thorough analysis of their experimental data. This figure was adapted from Wu et al. (2012)’s “Applications of Chromatography Hyphenated Techniques in the Field of Lignin Pyrolysis”.
Interestingly, an application of GC-MS is in fire investigations, whereby it can be used to determine the cause of a fire (and even whether it was deliberate or accidental!).
Although such incidents normally leave very little traces of evidence, scientists are usually able to use GC-MS to identify accelerants (i.e. substances that increase the speed of a reaction; e.g. kerosene that aids the spread of fire) which could be present in the crime scene. In particular, the chemical composition of the accelerant can provide clues about the manufacturer and thereby help the police to track down the arsonist.
Liquid chromatography-mass spectrometry (LC-MS) can be used to identify illicit drugs
Identifying unknown powders and liquids that could be illicit substances requires a presumptive test to be performed. This technique allows a broad classification of the mystery substance and can quickly determine if the sample is essential to the case (e.g. needs to be further analysed) within hours.
With that in mind, LC-MS is considered to be the gold standard for detecting and quantifying low concentrations of drugs in urine, human serum, and blood samples. In addition, this technique can also greatly reduce the cost and time spent on confirmatory tests.
What are the advances in mass spectrometry?
Despite all the wonderful technological advances and convenience that mass spectrometry has given us, there are still various factors that have highlighted its limitations.
For instance, there has been a rapid emergence of novel synthetic drugs that are unusual and often mixed with other street drugs. So, they are usually hard to identify and characterize. Besides that, since the frequency of drug abuse cases are on the rise, there is a demand for rapid methods that can handle the simultaneous analysis of many different compounds (this can be exceptionally challenging if we were to be dealing with novel or rare compounds). Therefore, it is imperative for us to continue innovating more comprehensive routine screening methods tailored for the analysis of relatively unusual substances.
A modern approach is the use of portable mass spectrometry devices that could enable the screening, collection of samples, and possible detection of trace chemicals within a sample present right at the crime scene. This could thus help researchers to speed up the investigation and reduce the workload imposed on forensic laboratories. However, the associated drawbacks include lower quality resolution as well as reduced detection limits.
DNA fingerprinting - another great tool for detectives!
We can also differentiate between different people based on their DNA fingerprint, which is a set of variable markers that are unique to each individual.
In particular, short tandem repeats (STRs) are highly variable repeated units that are up to six nucleotides long, with each individual having a distinct number of repeats (see Figure 2). Such variation mainly occurs in the non-coding regions of our genome since this will not disrupt the function and structure of proteins (and therefore have no effect on a person’s health).
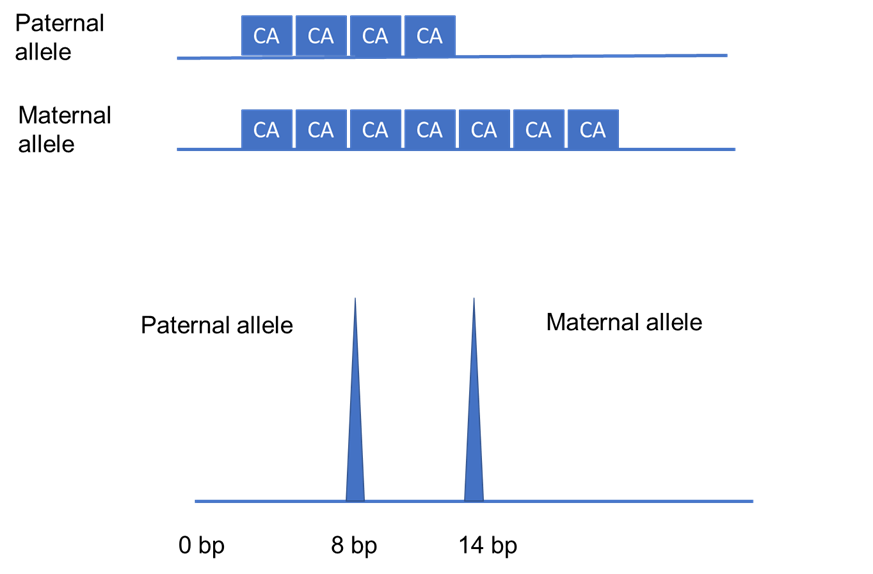
Figure 2: A schematic of an STR region of a heterozygous individual with repeated units of two nucleotides. STRs are inherited in a Mendelian fashion, that is - each copy of a gene (known as an allele) or region of the genome is inherited from either of the parents. As shown, the father passed down four CA repeats (equivalent to eight base pairs), whereas the mother gave seven repeats (which means 14 base pairs) to their child. This gives rise to a heterozygous individual who has a different number of CA repeats at a specific region of their genome. This can be seen as two separate peaks as the length of the STR repeats are different. All in all, this essentially demonstrates that the vast variations in heterozygosity and length of STRs between individuals make it very unlikely for two people to have exactly the same length of repeats for all STR regions tested. This figure was adapted from Saad (2005)’s “Discovery, development, and current applications of DNA identity testing”.
Usually, 13 of such regions are used to profile suspects and serve as conclusive evidence if there is a 100% match with the sample from the crime scene.
What does the experimental procedure involve?
DNA samples are first extracted from the crime scene and from the suspect themselves. Even a few cells from biological samples (e.g. saliva, hair, or fingernail clippings) is already plentiful as the technique involves making multiple copies of the repeat regions. In fact, only one nanogram of the sample is needed!
Next, DNA must be preserved and stored under cool conditions to avoid any sample contamination or fragmentation by UV radiation, extreme pH, and/or exposure to high salt concentrations. This is important as accurate data from many STR regions cannot be obtained with degraded DNA, and these could increase the probability of researchers assigning an incorrect match.
Moving on, short complementary fragments to the sample DNA called primers are used alongside different fluorophores that are specific for each STR region. In particular, primers mark the starting points for DNA amplification. The amplified fragments are then passed through a laser that excites the electrons of the fluorophores, whereby the fluorophores emit the absorbed light once the excited electrons return to their lower energy levels.
As a result, these can be seen as coloured peaks and the fragments can be separated by their molecular size. Using those, the STR profiles of the two samples can then be compared.
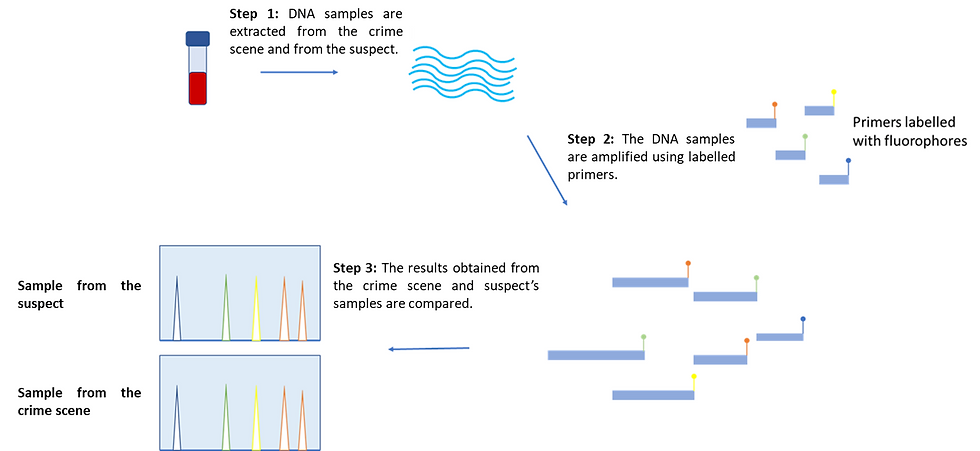
Figure 3: An overview of the DNA fingerprinting workflow. The experiments are performed on the suspect and the crime scene sample separately. To begin with, DNA is extracted from both samples, which is then amplified by using labelled primers that are elongated by DNA polymerase. Multiple copies are made for each labelled STR fragment. The fragments are then visualized and the two samples are compared. In essence, if the two profiles show a 100% match in all STR peaks, the suspect is likely involved in the crime. This figure was adapted from yourgenome (2020)’s “What is a DNA fingerprint?”.
The ethical issues of DNA databases
In the UK, the DNA profiles of convicted criminals, as well as suspects proven to be innocent, are collected and stored in DNA databases. Even though the DNA fingerprinting technique has proven to be highly effective in forensic investigations, there has been some debate regarding the ethical issues surrounding the establishment of DNA databases.
One notable problem is that DNA databases tend to be biased against certain ethnic groups such as Black and Latino individuals, whereby both of which tend to be disproportionately overrepresented in police records. Therefore, there is a higher chance that an innocent person from a minority background will get listed as a close match and become a suspect.
Apart from that, human error can also be an issue. For instance, if the scientist were to be given the context of the crime, this might influence how they interpret a result and, consequently, the conclusions they make. Thus, this showcases the biasing effect of contextual information when interpreting an experimental result.
Furthermore, most DNA profiles in databases are associated with relatively minor offenses (e.g. theft), whilst extreme cases (such as murder and rape) constitute up to only ~3% of the DNA profiles. As a result, this reduces the effectiveness of DNA fingerprinting and profile matching for more serious crimes.
What advancements are there on the horizon?
Contrary to how the DNA fingerprinting technique is portrayed on crime scene investigation TV shows, getting the results (even with the most advanced instruments) takes at least two hours. Nonetheless, the good news is that STR analysis can be combined with next-generation sequencing to speed up the analysis process (in which up to ten STR regions can be screened for simultaneously). This, therefore, allows for the screening of more STRs and it increases the accuracy of DNA profiling.
Another challenge that researchers ought to resolve is how they could analyse low-quality and contaminated DNA that is often collected from crime scenes. For instance, most DNA samples obtained from handguns or clothes contain DNA from more than one person. This essentially complicates and changes the STR profiles identified.
Henceforth, forensic scientists often use statistical computing systems to calculate the likelihood of these two samples coming from the same person. For example, TrueAllele can determine the genotypes of up to ten people in the input sample and assign probabilities to identify the real culprit.
The marvels of epigenetic fingerprinting
The human genome contains around 20,000 genes encoding important features, all of which are marked by cytosine and guanine rich regions called CpG islands.
CpG islands are located near the regions where transcription starts. When these regions are densely methylated (i.e. a methyl group is attached to the structure), proteins are recruited (and this creates a “transcriptionally repressive” state).
As a result, chromatin becomes more compact and the gene switches off. Given that different tissues have different functions, they each harbour a distinct set of genes that get switched on and off at certain time periods. This, therefore, creates a methylation pattern.
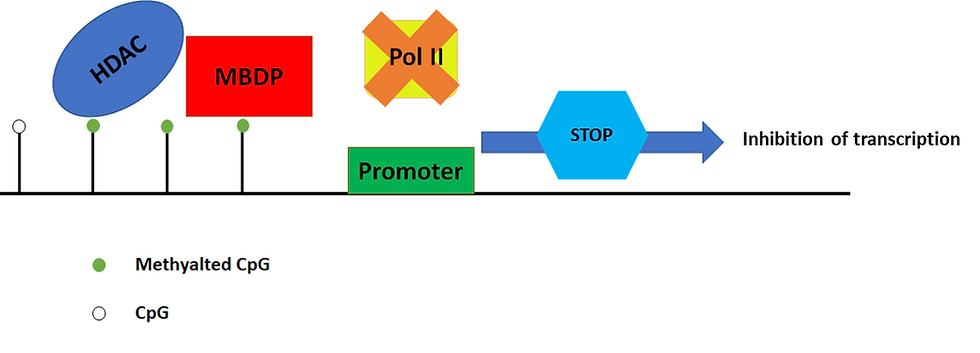
Figure 4: Methylated CpG islands induce transcriptionally repressive chromatin. The inhibition of transcription can be mediated by the binding of methyl-CpG-binding domain proteins (MBDPs) to the methylated regions, and they in turn recruit histone deacetylases (HDAC). HDACs are proteins that make DNA wrap itself more tightly and thus become less accessible to RNA polymerase (Pol II). As a result, the gene does not get transcribed. This figure was adapted from Sarkar et al. (2013)’s “Cancer development, progression, and therapy: an epigenetic overview”.
For forensic analysis, a few methylation sites associated with visible traits, such as hair and skin color, have been identified. These can then be used to determine the tissues of a sample from a crime scene. Meanwhile, although there are other epigenetic markers available for forensic analysis, DNA methylation is still preferred as it is stable enough to for in vitro (i.e. in test tubes) work.
What makes this technique effective?
The methylation pattern of DNA can provide important clues about the type of tissue, a person’s age, and gender (just to name a few).
As opposed to DNA profiling, methylation profiles are non-comparative - this means that the DNA from an unknown perpetrator is not a hindrance to the investigation, because the epigenetic profiles can help characterize and pinpoint a group of potential suspects.
Another advantage of this technique is its ability to distinguish between identical twins sharing the same DNA. The reason for this is because epigenetic markers are not strictly passed down to the next generation and are highly influenced by the environment. Therefore, methylated DNA sites can vary even between monozygotic twins.
DNA methylation microarrays - how do they work?
Forensic epigenomics has the potential to predict the characteristics of the perpetrator once large-scale DNA methylation analysis becomes technically feasible. This requires techniques that can handle lower-quality and reduced amounts of DNA as well as be able to analyse a large number of epigenetic markers simultaneously.
The commonly used technique in forensic epigenomics is DNA methylation microarrays (a.k.a. a DNA chip), in which a major advantage of using microarrays is that they are cost-effective and less labour-intensive than other high-throughput and sensitive techniques.
This technique involves screening the methylated regions of a sample by introducing it to a microarray of CpG island fragments, whereby the sample DNA binds to the complementary DNA on the microarray.
To begin with, the DNA samples from the crime scene are sheared into small 500 base pair (bp) fragments. The same workflow applies to the suspect’s sample. These sheared samples are then treated with enzymes such as HpaII (a widely used restriction endonuclease that can digest unmethylated fragments), leaving the methylated fragments unaffected. After that, the methylated DNA is amplified, denatured, and labelled with fluorescent markers. Once that is done, equal amounts of suspect’s and crime scene samples are mixed and added to the microarray, which is a glass slide consisting of spots that each contain different CpG islands.
Finally, after all of that, the scientist will perform some computer analysis to determine which of the three outcomes is obtained from this experiment (see Figure 5).
Red fluorescence indicates hypermethylation - this means that there is more methylation in the test sample in comparison to the suspect’s sample.
Green fluorescence indicates hypomethylation - this means that there is less methylation in the test sample when compared to the suspect’s sample.
Yellow fluorescence signals that there are identical methylation levels between both samples.
In summary, microarrays can identify CpG regions that have different degrees of methylation. This thereby helps forensic scientists to build a methylation profile of the unknown sample and characterize a possible suspect.
For example, if the crime scene sample shows yellow signals for markers of a particular trait (e.g. brown hair), this indicates that the perpetrator might have brown hair as well.

Figure 5: An overview of a microarray analysis workflow. DNA fragments representing the whole genome are fixed onto a glass slide, with each spot containing millions of copies of a single specific sequence. The fluorescently-labelled mixture of single-stranded DNA from the crime scene and the suspect are introduced. This is followed by using a laser scan to excite fluorescent labels, thereby allowing one to visualize the abundance of fragments in each spot. This figure was adapted from Mlakar & Glavac (2007)’s “DNA microarrays and their use in dermatology”.
What are the limitations of epigenetic fingerprinting and how can it be improved?
There are many limitations of forensic epigenetics that make it difficult to be applied to many investigations.
Firstly, large amounts of high-quality DNA is needed for microarrays if we were to carry out multiple experiments for multiple epigenetic markers. Since the DNA obtained from crime scenes are often in the picogram range, this necessitates the development of more sensitive technologies that can detect minute variation in methylation. In addition, there are only a few epigenetic markers that are currently available for use in investigations. Hence, this does not provide sufficient information for researchers to build an epigenetic fingerprint that is reliable enough to be used as a reference point.
Other than that, the available methylation patterns used to predict the visible traits of an individual are very limited and not very accurate because they overlap a lot. Therefore, we would need new types of biomarkers to help us better predict the visible traits of the suspect.
Some final thoughts
Based on what we have discussed, current biochemical techniques have been proven to be efficient and highly accurate at interpreting biological evidence. However, moving forward, we also need to view the problems from the perspective of citizens especially when it involves the use of personal information such as DNA fingerprint databases.
As a result, circumstantial evidence ought to be used in combination with biological evidence in order for us to paint a more accurate picture of the crime. This consequently helps to prevent the frequency of wrongful convictions in the future.
Author
Huong Giang Nguyen
BSc Biochemistry
Imperial College London
