Transposable Elements: A Double-Edged Sword
- Huong Giang Nguyen

- Mar 2, 2021
- 10 min read
Updated: Jul 6, 2023
Transposable elements, or transposons, are mobile genetic sequences that have the unique ability to “jump out” of a specific location in the genome and reinsert themselves elsewhere.
As a result, they are often also known as “jumping genes”.
Transposons have actually become an increasingly important research topic because the movement of such elements has proven to create both beneficial and harmful mutations at the site of insertion.
In particular, the uncovering of these genetic sequences was a groundbreaking discovery in the 1940s as it disproved the widely accepted assumption that genomes and genes are uniform in eukaryotes. Besides that, our knowledge of transposons and their roles (particularly in disease and the evolution of the human race) has massively expanded since their discovery and is, therefore, an important scientific avenue for research.
In this article, we will explore the dynamic relationship between the genome and transposable elements. In addition to this, we will also touch upon their use as genetic tools, which could be a promising endeavour to treat hereditary diseases in the (hopefully near) future.
What are transposons?
Intriguingly, about 46% of the human genome is occupied by transposable elements.
Having many active transposons commonly result in the development of genetic diseases as mutations can be caused by the “jumping in'' events. As a result, transposons are often referred to as “selfish” or “junk” DNA as they do not normally encode specific genes that benefit the host. Specifically, they encode enzymes that direct their own movement, i.e. the copying and inserting mechanism.
Though, bear in mind that this classification is widely debated as on the other hand, there is much argument that transposons are beneficial to the host because they have not been lost during evolution.
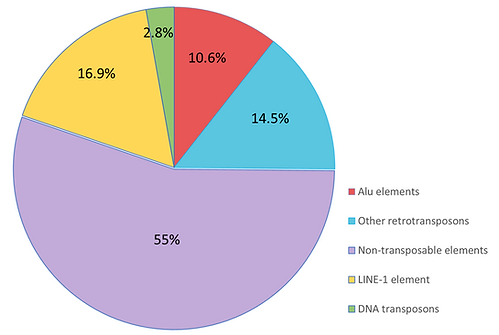
Figure 1: A pie chart showing the proportion of transposable elements in the human genome. Transposons (yellow, green, red, and blue regions) represent almost half of the total genome and are present in all eukaryotes. On that note, long interspersed nuclear element 1 (LINE1) is the most abundant transposon in humans. Meanwhile, Alu elements are a type of short interspersed elements, and they are the second most prevalent type of transposon in the human genome. Not to mention, they are also unique to primates. This figure was adapted from Cordaux & Batzer (2009)’s “The impact of retrotransposons on human genome evolution”.
How were transposons discovered?
Transposable elements were first characterised by a female scientist known as Barbara McClintock, whereby she associated the uneven pigmentation of corn kernels with transposons jumping in and out of genes. These transposons, in turn, influence the colour of the kernels.
To dive into greater detail, McClintock noticed that chromosomes suffered breakages at the same location due to the frequent insertion of a transposon. This led to her work contradicting the dominating view that genes are stable and that chromosomes do not go through rearrangements.
Moving forward, two major classes of transposable elements have been discovered since then, and they are RNA and DNA transposons.
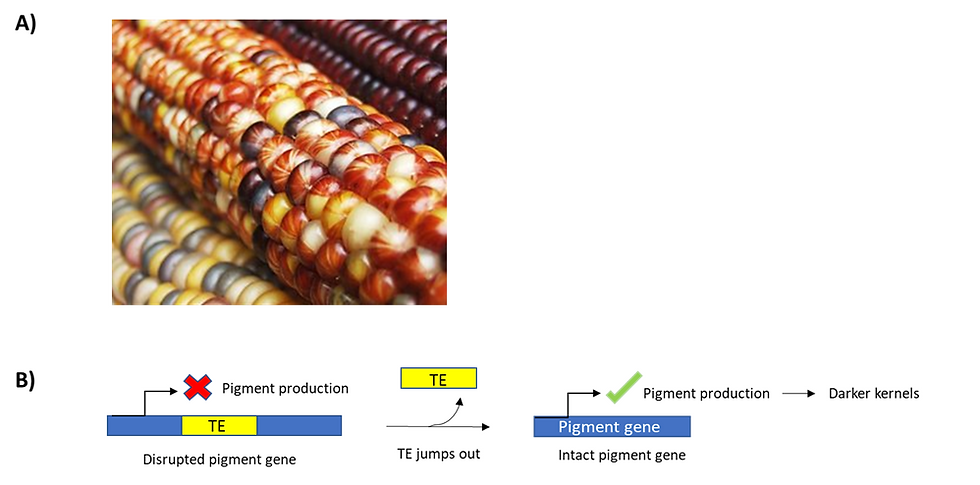
Figure 2: Corn kernels show a huge variation in colour and pattern due to transposition events. (a) An image of a mottling corn kernel. To provide some context, mottling is defined as the uneven pigmentation caused by the activity of transposons, which consequently disrupts the production of pigment in some kernels. (b) A schematic showing how transposon activity drives the mottling of corn kernels. Transposition can occur at multiple locations in pigmentation genes. In essence, if a transposable element gets nestled into the pigment gene, this disrupts the expression of pigment proteins, leading to yellow kernels. On the other hand, if transposable elements jump out the pigment gene, the pigment gene is then fully expressed. As a result, we would get pigmented, darker kernels. This figure was adapted from Feschotte et al. (2002)’s “Plant transposable elements: where genetics meets genomics”.
What kinds of transposons are there?
DNA transposons: the simplest type of transposable element
DNA transposable elements are essentially DNA sequences that can directly remove themselves from one location and reinsert themselves into another place.
The mechanism it undergoes to accomplish that is a relatively simple one compared to that of RNA transposons. This is because DNA transposons rely on only one type of enzyme in order to be able to move about, and that enzyme is called transposase.
Transposase, encoded within the transposon, uses a cut-and-paste mechanism to recognize and bind to sequences termed “inverted terminal repeats” (TIR). This eventually allows the enzyme to cut the transposable element out of the genome. Moving forward, the transposase is also responsible for cleaving the site where the transposable element is targeted to allow its re-integration into a new location within the genome.
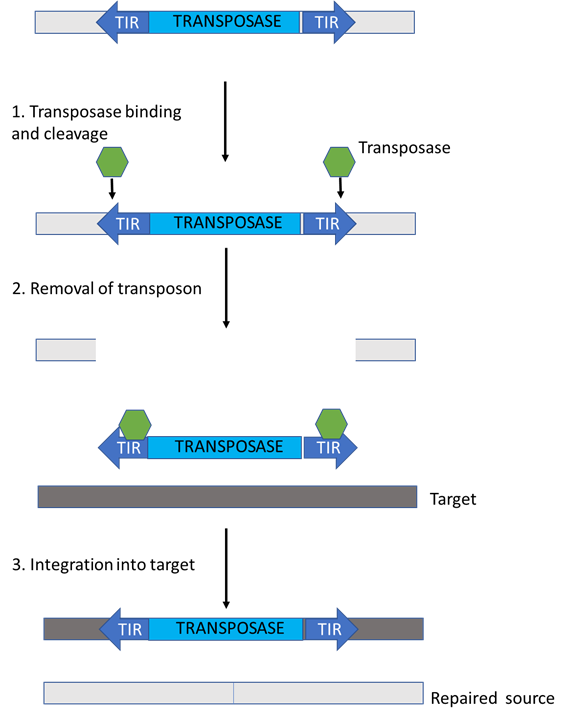
Figure 3: The structure and movement of DNA transposons. The transposase enzyme is encoded between two terminal inverted repeats (TIRs). After the transposase is expressed, the enzymes (green) bind to the TIRs and make cuts on either side of the transposon, thereby removing it. The DNA transposon can now move freely and it gets “pasted” into a different location directed by the transposase that is bound to it. On a lighter note, you can kind of imagine this process as a bit of a “cut-and-paste” process. This figure was adapted from Levin & Moran (2011)’s ”Dynamic interactions between transposable elements and their hosts”.
RNA transposons: the most successful transposable element
In contrast to DNA transposons, RNA transposons need an extra step during the process in order to convert themselves to DNA.
The simple reason for that is because they cannot jump back into the genome in their RNA form. Instead, they have to copy themselves from the genome, form mRNA, which is converted back into DNA that can integrate elsewhere in the genome. So basically, the original version of the transposon remains intact and a copy is integrated into a new location, resulting in an abundance of these repetitive sequences throughout the genomes.
The most abundant RNA transposons include LINE-1 elements, which are thought to be the only active transposable elements in humans that can move on their own.
Other than that, Alu elements are also important to mention as they are the most successful transposons thanks to their role in evolution and diversity due to frequent insertion into different genes. However, since they have lost their ability to move, they have to depend on LINE-1 for transposition.

Figure 4: A diagram showing the structure of RNA transposons and how they move about within the genome. The LINE-1 element contains two open reading frames (ORFs). The ORF is a segment of a gene that encodes amino acids and is translated into protein without stop codons. Besides them, LINE-1 also contains a 5’ untranslated region (5’ UTR) that binds RNA polymerase so that the ORFs can be transcribed. ORF1 and ORF2 are then translated into protein. The ORF2 protein has an active role in directing the movement once they bind the mRNA form of the transposable element (with the help of ORF1 proteins). In particular, ORF2 has endonuclease activity, which means that it can cleave the target DNA within the genome. Besides that, it also has reverse transcriptase activity, meaning that it can make DNA from mRNA. As a result, the translated ORF2 can convert transposable element mRNAs into DNA and cleave a new target location allowing the transposable element DNA to be inserted. This figure was adapted from Levin & Moran (2011)’s ”Dynamic interactions between transposable elements and their hosts” and Ostertag & Kazazian (2005)’s “Retrotransposition and Human Disorders”.
Overactive transposons cause diseases
You might ask - what happens when these mobile sequences go rogue and start to move about uncontrollably as they like within the genome?
Well, this essentially translates to bad news for the host in the short term. And that’s because this event causes local and more widespread mutations, which could lead to undesirable changes in gene expression and thereby cause diseases.
To begin with, local changes include the insertion of transposons into promoters, i.e. the DNA located in front of genes where transcription is initiated, which can lead to the overexpression of genes. This, essentially, is the hallmark of many variants of cancer.
On the other hand, large-scale structural changes in chromosomes such as deletions (i.e. removal of large sequences of DNA) are quite frequent. As a result, mutations arising from the removal of DNA transposons can lead to the disruption of promoters and lowered levels of gene expression. For example, if a gene important in suppressing tumours is poorly expressed, that will allow tumour cells to proliferate uncontrollably. In short, terrible news as well.
And not to mention, there are 65 documented genetic diseases that can be caused by the insertion of transposable elements within egg and sperm cells. This, in turn, leads to the development of hereditary conditions, whereby examples of such diseases include breast cancer and a blood coagulation disease known as haemophilia.
How does the host fight against transposon activity?
As you probably would have figured out at this point, it can be difficult for the host to survive persistent insertion events, particularly if there has been an accumulation of mutations within vital genes.
Nonetheless, the good news is that many organisms have evolved mechanisms such as methylating their DNA to protect themselves against such events. Methylation is a gene-inactivating process whereby a small molecule tag (a methyl group) is deposited onto the guanine nucleotides. This ultimately leads to the DNA becoming tightly wrapped and inaccessible to the transcription machinery.
Meanwhile, just like most promoters, the promoter region of LINE-1 also contains CG-rich regions. These are basically DNA sequences possessing extended repeats of cytosine and guanine in front of their genes, and they play a big role in determining whether the gene is expressed or not. Crucially, they are known to be methylated when a gene needs to be repressed.

Figure 5: A simple illustration of how methylation of CG-rich regions control transcription. Promoters often contain CG-rich stretches that can be unmethylated, which allows transcription proteins such as RNA polymerase to access the promoter and the gene located next to it, thereby leading to the production of mRNA. On the other hand, if the promoter is methylated, that causes the tight wrapping of the DNA. Therefore, transcription proteins will not be able to help transcribe the gene.
Normally, transposon promoters are normally highly methylated and thus are generally transcriptionally silenced. However, one of the interesting (and frustrating) parts about cancer cells is that they frequently demethylate transposons. Hence, they end up reactivating the transcription of LINE-1, which consequently directs the reinsertion of copied LINE-1 elements elsewhere. This thereby contributes to further genomic instability, which is a classical hallmark of cancer.
How do transposons drive evolution?
The high quantity of transposons (remember, 46% of our genome!) suggests they may have had a significant role throughout human evolution.
Interestingly, transposons have been shown to frequently insert themselves throughout the genome over the last millions of years!
It was even implied that transposons can be activated under stress because this can create new functions that may aid host survival. Or more specifically, if the insertion of transposons helped the host to survive (e.g., it increased the host’s ability to absorb nutrients), the insertion was inherited by future generations. In contrast, if an insertion resulted in the death of the host, the transposon will not be inherited. All in all, this is called natural selection.
Thus, over time, this may be an explanation as to why transposons become immobile in the long run as any new insertions would have had undesirable outcomes. It also explains why almost all transposons in humans are inactive (well, apart from a few that can occasionally cause disease).
How is human evolution impacted by transposons?
Meanwhile, transposons also have a major impact on primate and human evolution specifically. This is indicated by the frequency of transposition in the sperm and egg cells that allows the inheritance of such changes. In fact, there is about one insertion in every 20 births, which explains the variation in transposons among humans.
One of the processes that get accelerated by transposon insertions is the evolution of the human brain. This was made possible by the variation caused by random transposon insertions as well as natural selection.
In addition, Alu elements have also played an important role in shaping human evolution by altering the expression of genes that are close to the insertion sites. Since they are the most abundant transposons in the human genome, Alu elements might help to explain the variation we see today within the human race. Hence, identifying such elements can be a useful way for us to study the evolution of the human genome and different races.
Other than that, the genetic diversity induced by transposons, in general, could shed light on why some people are at increased risk of certain diseases.
Still, it is important to note that the positive effects of transposons on genome evolution is likely a by-product rather than the key reason for their evolutionary success. In short, the fact that transposons often cause disease in humans is the main evidence supporting this viewpoint.
Transposons as new tools for molecular surgery
Despite their potentially disruptive nature, the transposons’ ability to integrate randomly in the genome makes them useful at inactivating genes as well as introducing new genes.
Fortunately, the transposition process can be controlled if the transposase and gene of interest flanked by Terminal Inverted Repeats (TIRs) are expressed from different pieces of circular DNA called vectors. As a result, the gene of interest can only be removed and reinserted elsewhere if the transposase is added to the target cells.
As a matter of fact, if we were to apply our knowledge in this to solve a specific problem, this can be used to restore the expression of a protein that has been lost due to a mutation. This itself is prevalent in diseases such as sickle cell anaemia, which can be treated if the sequence of the missing enzyme is re-inserted.
Besides that, this set-up can be used to introduce any gene into the genome and has been widely used in experimental models such as fruit flies to show the potential of transposons in treating genetic diseases. However, although this method has been successful in combating diseases, the location of the insertion is still entirely random. Hence, vital genes can still be disrupted.
But overall, this effectively showcases how transposons could have tremendous potential in the field of gene therapy.
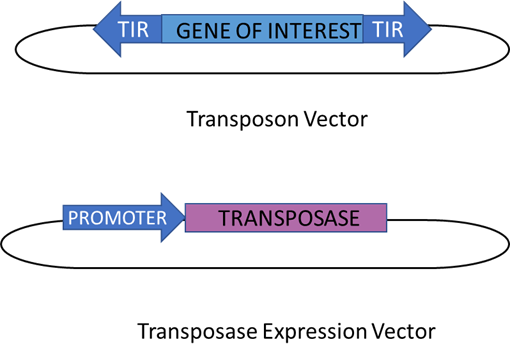
Figure 6: The gene transfer system using DNA transposons. This system consists of two components - a transposase gene and a separate transposable element in which the transposase gets replaced by a gene of interest. On each end of the gene, TIRs can be found where the transposase can cut to allow the removal of the element, and this happens the same way as discussed before for traditional DNA transposons. As a result, the transposase gene is expressed from a strong promoter from a different vector. Meanwhile, the drawback potential of this system is that the transposon could be accidentally inserted into important genes. This could result in its vital functions being disrupted and the host developing a genetic disease. This figure was adapted from Muñoz-López & García-Pérez (2010)’s “DNA Transposons: Nature and Applications in Genomics”.
Why is it important to study transposable elements?
In general, transposable elements are frequently involved in regulating the expression of genes because certain transposons encode the binding sites for transcription factors.
On an evolutionary scale, transposons can distribute binding sites for transcription factors, which in turn, can drive the evolution of different species by giving rise to new traits.
Besides all of that, transposons are an intriguing area of research - not only as targets for therapeutics but also for studying the evolution of humans. Who can really tell - perhaps future studies could shed some light on how the differences among people are the product of a transposon-driven evolution? Or maybe, we might be able to finally explain how certain people could fight off certain diseases better than others.
Author
Huong Giang Nguyen
BSc Biochemistry
Imperial College London
#transposons #evolution #genetics #disease



